Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Uday Bikash Oraon* and Jayanta Tarafdar
Received: March 30, 2018; Published: April 11, 2018
*Corresponding author: Uday Bikash Oraon, Department of Plant pathology, BCKV, Mohanpur, Nadia, West Bengal, India, 741252
DOI: 10.26717/BJSTR.2018.03.000948
Chilli (Capsicum annum L) is popularly known as 'wonder spice' and is an important cash crop and remunerative vegetable crop of India and is grown both for home market and export. Chilli leaf curl complex disease is one of the major limiting factors in chilli production & the reasons behind this complex disease are whitefly (Bemisia tabaci) transmitted begomoviruses, thrips & mites, causing significant reductions in yield and quality of chilli. In our present investigation attempt was taken to study the incidence and distribution of the Chilli Leaf Curl Complex Disease predominantly occurring & infecting chilli crops in different locations of West Bengal & detection of begomoviruses through PCR based method using one pair of degenerate primer SPG1/SPG2. The survey was carried out in 18 villages and total 54 fields in six districts randomly selected in West Bengal during July- August, 2015 when the crop was at 2-3 month old. The highest leaf curl incidence was noticed in Cooch Behar district (30-90%) followed by Burdwan (20-85%) and North 24 parganas (45-75%) in 2015. While the lowest incidence was observed at Purulia (18-43%) district. Among six districts Cooch Behar, Jalpaiguri (Madarihat), Burdwan, North 24parganas, Nadia chilli leaf curl samples showed positive reactions in PCR amplification of a target sequence (920 nt long) specific to begomovirus confirmed the presence of begomovirus in the samples tested. Moreover, these studies supply precise information about the frequency and distribution of distinct begomoviruses in chilli in West Bengal.
Keywords: Survey; Chilli leaf curl; Begomovirus; PCR; West Bengal
Abbreviations: CLCV: Chilli Leaf Curl Virus; CTAB: Cetyl Trimethyl Ammonium Bromide; PCR: Polymerase Chain Reaction; NB: North Bengal; SB: South Bengal
At present, chilly is produced in India about 1260.1 thousands metric ton from an area of 792.1 thousands hectare [1]. India is the largest producer, consumer and exporter of chilli, exporting to USA, Canada, UK, Saudi Arabia, Singapore, Malaysia, Germany and many countries across the world and contributes 25% of total world production. Andhra Pradesh ranks first in area, production and productivity of chilli contributing 30% of the total area and 51% of total production and West Bengal share is 5% only. Agricultural crops of India, like other countries, are also threatened by several biotic and abiotic factors [2]. Chilli leaf curl complex disease is one of the major limiting factors in chilli production caused by whitefly (Bemisia tabaci) transmitted begomoviruses, thrips & mites, causing significant reductions in yield and quality of chilli. Out of the 22 viruses infecting chilli, chilli leaf curl virus (CLCV) is one of the major limiting constrain for chilli production in the Indian subcontinent and is consistently caused by begomoviruses [3-5].
Leaf curl disease of chilli showed typical symptoms of begomoviruses (like upward leaf-curling, distortion, vein yellowing or more generalized leaf yellowing and/or stunted growth with bushy appearance of chilli plants) and these symptoms result heavy economic losses of about 15 billion US Dollar per annum worldwide [6]. The genus Begomovirus is the largest genus of family Geminiviridae & contains approximately 300 species [7], only infects dicotyledonous plants and is transmitted by the phloem feeding whitefly, Bemisia tabaci (Genn.). Chilli leaf curl etiology was established during the 1960s in India [8,9]. In 2004, epidemics of CLCD occurred in Jodhpur, the main chilli-growing area in Rajasthan, India and then Senanayake et al. [5] have reported first time chilli leaf curl virus on chilli crop from Jodhpur (Rajasthan). Recently identification of Tomato Leaf Curl Virus (ToLCV) strain causing leaf curl disease in Tomato and Chilli in Maharastra reported by Chavan et al. [10], results in 90-100% yield loss. The rapid evolution of virus variants, the massive increase of vector populations (with the appearance of whitefly 'B' biotype) and the introduction of modem human agricultural practices have contributed to a rapid and worldwide spread of the virus [11]. However, there are no published reports on the distribution of chilli leaf curl complex disease in major chilli growing areas of West Bengal although, it is important disease of chilli crop.
The single point method was adapted to the survey the chilli leaf curl disease incidence at a critical period of crop growth stages in six districts namely Coochbehar, Jalpaiguri, Nadia, Purulia, North 24 Parganas and Burdwan. From each district one block selected based on chilli cultivation status & number of village surveyed. The survey was confined to two or three villages of each block and villages were selected at random. These survey villages were visited for once when the crop was at 2-3 month old during July- August, 2015. Data on the total number of plants, number of virus infected plants in per square meter. The disease diagnosis in the field was based on typical symptoms of begomoviruses, mites & thrips infection. The percent disease incidence was recorded at random in different locations at per square meter in the field by counting total no of plants and no. of plants showing chilli leaf curl symptoms using the formula [12] given below- ,
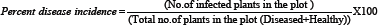
Chilli plants showing upward (abaxial) leaf-curling, distortion, vein yellowing or more generalized leaf yellowing and/or stunted growth with bushy appearance were randomly selected and samples (healthy & infected) collected from six different districts of West Bengal.
Total DNA were extracted from 100 mg of infected and healthy plants using Cetyl trimethyl ammonium bromide (CTAB) method [13] and modified by Sharma et al. [14] was followed for extraction of DNA. Presence of begomovirus was confirmed by PCR amplification with a begomovirus specific degenerate primer pairs SPG1/SPG2 (5’-CCCCKGTGCGWRAATCCAT-3y5- ATCCVAAYWTYCAGGGAGCTAA-3' [15], which amplify replication associated protein (AC1) gene partial segment. The PCR amplification was carried out in a thermal cycler (Mastercycler, Eppendorf, AG 22331, Germany). The PCR reaction mixture contained 2μl DNA sample, 1μl of each primers (Conc. 10mM), 1μl dNTPs (10 mM), 1.5 μl Mgcl2 (25mM), 2.5μl 10x reaction buffer, 0.5μl Taq polymerase and 15.5μl sterile HPLC-H2O, that will give rise to a volume of 25μl PCR reaction. The conditions for PCR reaction were, an initial denaturation at 940C for 5.00minute followed by 34 cycles of denaturation at 940C for 30sec., annealing at 590C for 30sec and extension for 1minute at 720C. Then final extension at 720C for 7 minute was included. Amplified PCR product were separated by electrophoresis on 1.5% agarose gel and DNA fragments were visualized using ethidium bromide strain, here 1 kb DNA ladder (Fermentas, Life Science) is used & documented by Gel documentation system (Vilber-Lormout, France). The expected amplicon of 920bp were obtained from the DNA of five diseased leaf samples chosen out of seven. The amplified products were stored in-40C for further use.
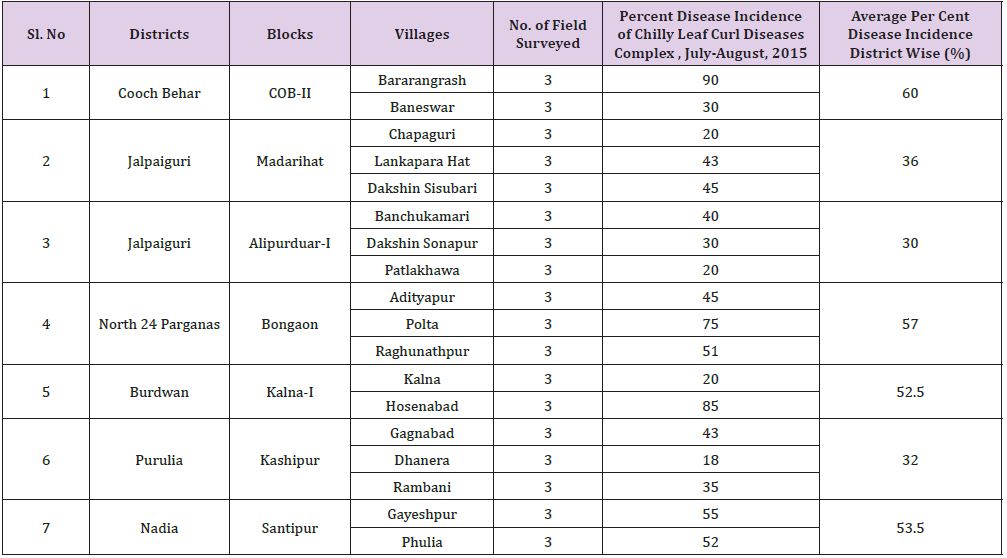
Field survey were conducted during July-August, 2015 in 18 villages and total 54 fields in major chilli growing areas of six districts namely Coochbehar, Jalpaiguri, Nadia, Purulia, North 24 Parganas and Burdwan. The results of survey revealed that most of the fields showed mixed infection of virus, mites and thrips. Different kinds of disease symptoms like mild to severe leaf curling (downward and upward), leaf blistering, leaf crinkling and leaf narrowing of leaves with stunted growth and bushy appearance were observed in the field conditions (Figure 1). The severely infected plants produced no fruit; however, less infected plants produced fruits of significantly reduced sizes. The disease incidence was found to be varied from location to location and region to region. Disease incidence of about 20-90% was found in North Bengal (NB) and it was recorded as 18-85% in South Bengal (SB) (Table 2).
Figure 1: Variable Symptoms of Chilli Leaf Curl Produced at (a) Cooch Behar (b & c) Jalpaiguri (d) N 24p (e) Burdwan (f) Nadia (g) Purulia.

The highest leaf curl incidence was noticed in Bararangrash (90%) followed by Hosenabad (85%) and Polta (75%) whereas the lowest leaf curl incidence was noticed in Dhanera (18%) (Table 1). When the leaf curl disease incidence data was analyzed at district wise the average maximum incidence was found 60% at Cooch Behar, while the least incidence was observed at Jalpaiguri (30%) may be due to favorable environmental condition was found for the growth and transmission of white fly (Bemisia tabaci), mites and thrips (Table 1) and (Figure 2). Infection of leaf curling was found moderate to severe in all blocks. Whereas downward curling and crinkling of leaves giving an inverted boat shaped appearance of mite infection was severe in Alipurduar-I and upward leaf curling with elongated petiole indicated thrips abundance in Kashipur.
Figure 2: Minimum & Maximum Chilli Leaf Curl Incidence in Six Districts of West Bengal.
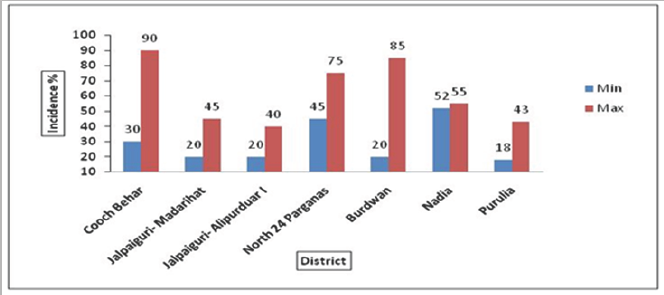
Table 1: Survey of Chilli Leaf Curl Complex Disease Incidence during the Growing Period of 2015 in Different Villages of West Bengal (WB).
Samples which showed distinct visible symptoms of begomovirus infection were subjected to polymerase chain reaction (PCR). One set of universal degenerate primers (SPG1/ SPG2) used for the amplification and confirmation of the partial replication associated protein gene (AC1) with expected DNA fragment of 920 bp from the virus infected samples. Positive reactions in PCR amplification of a target sequence (920 nt long) specific to begomovirus confirmed the presence of begomovirus in the samples tested. But Alipurduar-I and Kashipur samples gave negative results on PCR and the symptoms produced on chilli crop indicating towards mite & thrips infestation respectively (Table 2) and (Figures 3 & 4).
Table 2: Types of Chilli Leaf Curl Complex Disease Symptoms and PCR Results of the Samples Collected from Different Blocks of West Bengal during Crop Growing Periods of 2015.

*Randomly One Isolate from each Location was taken for Molecular based Study.
Symptom codes: LC: Leaf Curl; ST: Stunting; Chl: Chlorosis; LR: Leaf Rolling, B: Boat Shaped Leaf; EP: Elongated Petiole; (+) = Presence of the Virus; (-) = Absence of the Virus.
Figure 3: PCR Amplification of Begomovirus Samples Collected from North Bengal (NB) using Begomovirus Specific Degenerate primer SPG1/SPG2. Lane M- 1kb DNA Ladder, 1- Madarihat Sample, 2- COB-II Sample, 3-Alipurduar-I sample.
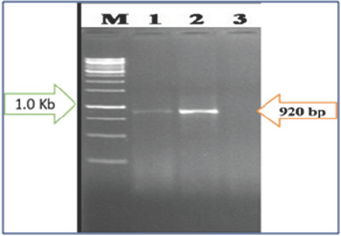
Figure 4: PCR Amplification of Begomovirus Samples Collected from South Bengal (SB) using Degenerate Primer Pair SPG1/SPG2 Sowing the size of Amplicon 920 bp. Lane M- 1kb DNA Ladder; 1: Bongaon, 2: Kalna-I, 3: Santipur, 4: Kashipur.
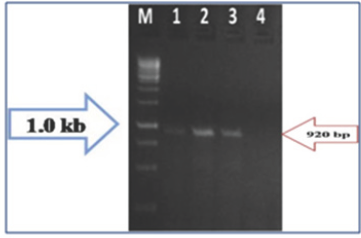
The PCR detection of begomovirus using degenerate primers confirmed the association of begomoviruses with chilli growing in West Bengal; it is due to cultivation of susceptible local cultivars prevailing in the districts. The variation of symptoms between different chilli growing areas indicated mixed infections of begomoviruses, thrips and mites. So the high frequency and distribution of begomoviruses in chilli throughout West Bengal could lead to epidemic in upcoming crop periods if proper management tactics are not taken into view.


