Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Carmen Socaciu*1, Corina Moraru1, Dan Eniu2, Adelina Staicu2, Claudiu Rachieru2, Rares Buiga2 and Andreea Iulia Socaciu2
Received: March 19, 2018; Published: April 10, 2018
Corresponding author: Carmen Socaciu, University of Medicine and Pharmacy "Iuliu Hafieganu” Proplanta Srl, str Trifoiului 12G, Cluj-Napoca, Romania
DOI: 10.26717/BJSTR.2018.03.000941
Lipidomics, a promising detection method, may serve as a novel prognostic approach for breast cancer patients. Using high performance liquid chromatography coupled with electrospray ionization triple quadrupole time of flight mass spectrometry, this pilot study aims to detect the serum lipidomic signature of patients with confirmed invasive ductal carcinoma, at three different stages (stage I—III) comparatively to healthy controls. Two different multivariate and univariate statistical models were applied for evaluating the lipid species, by PCA, PLS-DA, AUROC and T-test, as potential biomarkers in the diagnosis of this type of breast cancer. Our results revealed that lipid biomarkers responsible for the discrimination between breast cancer patients and healthy controls belong mainly to diacylglycerol and phosphatidylcholine derivatives of different unsaturated fatty acids.
The breast cancer evolution from stage I to III may be also predicted also by three diacylglycerols and one phosphatidylcholine derivative, while discrimination between pre-menopause and menopause patients can be predicted only by one potential biomarker, a lyso- phosphatidylcholine (18:2/0:0). These preliminary data are complementary to previous findings which reflect the key role of phospholipid metabolic pathways in breast cancer diagnosis and evolution, as well the signaling role of diacylglycerols as intracellular lipolysis markers. It is obvious that such data need validation for larger patient cohorts, to establish a significant number of relevant biomarkers for breast cancer diagnosis and prognosis.
Keywords: Breast Cancer; Lipidomics; UHPLC-ESI+-MS; Potential Biomarkers
Abbreviations: BC: Breast Cancer; HPLC: High-Performance Liquid Chromatography; MS: Mass Spectrometry; NMR: Nuclear Magnetic Resonance Spectroscopy; PLs: Phospholipids; ESI: Electro Spray Ionization; LPC: Lysophos Phatidyl Choline PC: Phosphatidyl Choline; PS: Phosphatidyl Serine; SM: Sphingo Myelin; PIs: Phosphatidyl Inositides; SFAs: Saturated Fatty Acids; GPs: Glycerophos Pholipids, GLs: Glycerol Lipids; SPs: Sphingolipids, FAs: Fatty Acids; STs: Sterol Lipids; PLSDA: Partial Least Square Discrimination Analysis; IDC: Intra Ductal Carcinoma; FMF: Find Molecular Features; ROC: Receiver Operating Characteristic; PCA: Principal Component Analyses; PLS-DA: Partial Least Square Discriminant Analysis; PG: Phosphatidyl Glycerol; FC: Fold Change; PKC: Protein Kinase C
Breast cancer (BC) is the main leading cause of cancer-related mortality in women, with prevalence above 25% [1]. The current non-invasive detection methods for diagnosis and surveillance include mammography, histopathology and blood tests (such as antigens and protein patterns). Those methods do no show a real success for a long-term survival rate since more sensitive biomarkers for even earlier detection along with molecular targets for better treatments are still expected [2,3]. Altered cellular functions of BC cells lead to uncontrolled cellular growth, with morphological and metabolic changes. Metabolomics and lipidomics are emerging approaches used to identify metabolic biomarkers of cancer, assisted by multivariate and univariate statistics, to understand the specific metabolic pathways and to monitor anticancer therapies [4,5]. Plasma or serum metabolome express the cellular activity and physiological or pathological cell-tissue relation, by modifying its "signature” [6-8].
BC is known to display an important heterogeneity due to hormonal status and other factors, from histology to prognosis,metastatic evolution and treatment responses. Therefore, considering the acute need for an early diagnosis, many studies explored the blood serum molecules as key biomarkers of diagnosis in BC [9-25]. Some lipid categories are intimately involved in the regulation of cell signaling, but still understudied. During malignant transformation, the lipid metabolism is altered, as it was yet investigated since two decades at the gene and protein expression level. However, recently lipidomics enabled the investigation of specific pathways where lipid metabolites may be involved [16-25]. The High-Performance Liquid Chromatography (HPLC) coupled with Mass spectrometry (MS) is one of the most appropriate analytical technologies which provide the separation, detection and identification of active metabolites from a tissue, body fluids or cell cultures by an untargeted or targeted approach.
The lipid profiles indicate the specific signature of BC, and may offer information regarding novel biomarkers for early detection and prognosis, or monitoring the efficacy of anticancer treatment. The comprehensive analysis of metabolites by 'omics' technology using HPLC-MS or nuclear magnetic resonance spectroscopy (NMR) are currently used to identify and define the metabolic phenotype of cells, tissues or biological biofluids [9-14]. Recently, there were identified different metabotypes of 18BC cell lines, by non-targeted metabolomics and more than 900 metabolites with extensive heterogeneity across metabolic pathways [15]. Higher plasma levels of lysophosphatidylcholine (LPC, 18:0), were related to a low BC risk [16] and similarly results were obtained by GC-MS, direct infusion MS or NMR [2]. Other comprehensive lipid profiling of plasma in BC patients compared with benign tumors revealed increased phosphatidylglycerol (36:3), phosphatidylinositol's PI (32:1,34:1,38:4) or glucosylceramide (d18:1/15:1) as novel biomarkers [17].
Phospholipids (PLs) are main constituents of cellular membranes and play important functional, structural and metabolic roles in cellular processes that undergo transformation, cancer progression, and metastasis. PLs are particularly suitable for HPLC- MS analyses involving electrospray ionization (ESI) techniques, which are well established in the proteomics field, have also been implemented in analyses of lipids [18,19]. When compared to the total PLs content, plasma phosphatidylcholine (PC) levels were reduced while LPC increased in aggressive cancer cells while phosphatidylserine (PS) levels remained unchanged. MS analysis showed alterations in the classes of PC, LPC, sphingomyelin (SM), and increases of phosphatidylinositides (PIs) with saturated fatty acids (SFAs), considered as signaling molecules. Nevertheless, detailed information about PLs changes in cancer progression is still lacking [20,21]. Around twenty five PLs were found to be statistically different in the serum of BC patients when compared with benign and healthy controls, twelve PLs being differentially expressed, six up-regulated and six down-regulated [21,22].
There were reported recently, new plasma lipid biomarkers for early diagnosis of BC, more than 840 lipid species being identified by UHPLC-QTOF-MS, mostly represented by glycerophospholipids (GPs), glycerolipids (GLs), sphingolipids (SPs), free fatty acids (FAs) and sterol lipids (STs). Using multivariate Partial Least Square Discrimination Analysis (PLSDA) model, six differentiating lipids were identified, namely PC (40:7) and (46:1), TG (26:1) and DAG (36:3) at higher levels in BC, around 20 lipid species being considered as putative serum biomarkers for BC diagnosis [24]. This pilot study aimed to assess the serum lipidomic profiles in Invasive Ductal Carcinoma, ER+ diagnosed BC patients, at different stages (from I to III), comparing with healthy patients, using a liquid chromatography-mass spectrometry (LC-ESI+-QTOF MS) platform (Bruker) lipidomics approach, considering that specific signatures and putative biomarkers can be detected in relation to the BC evolution and the alteration of phospholipid metabolism.
The protocol of this study was approved by "Iuliu Hatieganu” Medical University Ethics Committee. For each subject was obtained written informed consent, before entering the study. After securing the patients' agreement on the protocol, blood samples were collected from 31 patients (PS, mean age of 52.5±9.12 years old) with confirmed BC in three stages (3 in stage I, 17 in stage II and 11 in stage III) and 5 healthy controls (MS, mean age of 46.69±3.27 years old). Seventeen out of 31 patients from PS group, and four out of 5 from MS group were in post-menopause status. The diagnosis of BC was biopsy-confirmed to belong to invasive intraductal carcinoma (IDC) breast tumors, before any specific treatment, and classified in molecular subtypes using immunohistochemical staining.
Blood serum samples were collected by venipuncture into vacutainer tubes without anticoagulant. The blood serum was obtained by centrifugation at 2000 rpm for 10 minutes and aliquots of 1 ml were frozen and stored at -80 °C until analysis. The serum lipids were extracted using the standard Bligh and Dyer method slightly modified: to a volume of 0.2 ml serum, there was added 0.2ml methanol 95% to precipitate proteins. The mixture was vortexed for 20 s, kept at 4°C for 15min, then 3.5ml chloroform were added, again vortexed for 30s, followed by an addition of 0.2 ml water to induce phase separation. After mixing, the vials were centrifuged at 8000g for 10min and the lipid phase was collected and dried under Nitrogen. The dry residue was solved in a volume of 500μl mixture of Acetonitrile: Isopropanol: Water (65:30:5, v:v:v), sonicated 10min, filtered through 0.2μm PTFE filters and 5μl were injected in the HPLC column.
The HPLC- MS analysis was performed on a Bruker Daltonics MaXis Impact device including a Thermo Scientific HPLC UltiMate 3000 system with a Dionex Ultimate quaternary pump delivery and ESI+-QTOF-MS detection, on C18 reverse-phase column (Acclaim, Dionex) (5μm, 2.1 x 100 mm) at 55°C and a flow rate of 0.25ml/ min. The mobile phase was represented by a gradient of eluents A (water: acetonitrile, 6:4, containing 10mM ammonium formate) and B (isopropanol: acetonitrile, 9:1, containing 10mM ammonium formate). The gradient system consisted of 75% A: 25% B to 50% A: 50% B (min 0 - 4 min), to 3% A: 97% B (min 4-19min), isocratic (min 19-23) and return to 75% A: 25% B (min24 -28). The MS parameters were set for a mass range between 50-1000 Da. The nebulizing gas pressure was set at 2.8bar, the drying gas flow at 12 L/min, the drying gas temperature at 300 °C. Before each chromatographic run, a calibration with sodium formate was done. The control of the instrument and data processing used the specific software TofControl 3.2 and Data Analysis 4.2 (Bruker Daltonics).
Data Analysis 4.2 was used to obtain Find Molecular Features (FMF) followed by generation of an advanced bucket matrix, automatically obtained after time alignment and normalization. The data pre-processing, alignment, bucketing and normalization of FMF obtained were done with Profile Analysis 2.1. Only metabolites detected in more than 80% of the samples were included in the bucketing and statistical analysis. The multivariate statistics specific to untargeted metabolomics consisted initially in Principal Component Analysis (PCA-scores and loadings) delivered by Profile Analysis 2.1. Software and 44 molecules were selected for further analysis. The selected metabolites were further analyzed by online Metabo Analyst 3.0 platform (https://www.ncbi.nlm.nih.gov/pmc/ articles/PMC4489235), by multivariate analysis (PCA and PLS-DA) as well for the univariate analysis (calculation of P-values from T-test).
The same software was applied to build the Receiver Operating characteristic (ROC) curves and Areas under the ROC curve (AUROC) values, to evaluate the sensibility and selectivity of the potential biomarkers. Finally, the hierarchical clustering analysis using the statistical significant metabolites was performed, as a heatmap showing the positive or negative deviations of each sample and significant metabolite from the background. The relevant molecules were identified by searches based on m/z values and filtrations of possible serum metabolites from specialized databases like Lipid Maps (http://www.lipidmaps.org/tools/ms/LM SD_search_mass_ options.php) and Human Metabolome Database (http://www. hmdb.ca/spectra/ms/search).
Based on the raw data from HPLC chromatograms and the complex advanced bucket matrix (FMF) provided by the Profile Analysis Bruker instrument, after alignments, normalizations, the selection of 80% common molecules in the samples was complemented with MS data (m/z values from 100 to 1000). For an adequate interpretation, initially it was applied a non-targeted, unsupervised, multivariate statistics to differentiate the pathologic PS group against control, MS group and 44 compounds were selected for further analysis based on PCA loadings results. The analysis was followed by the unsupervised principal component analyses (PCA) and supervised partial least square discriminant analysis (PLS-DA) on the 44 molecules selected, using the online software Metabo Analyst 3.0. Figure 1 represents the 3D representation of PLS-DA scores for the first 3 components (PC1-PC2-PC3) to discriminate between PS and MS groups and also the PCA loadings, which shows the molecules which may be responsible for this discrimination (Figure 1).
Figure 1: Three dimensional representation of PLS-DA scores (left) for the first 3 components ( PC1-PC2-PC3) to discriminate between the BC patients (PS) and healthy controls (MS) groups and PCA loadings (right) which shows the molecules mainly responsible for the discrimination.
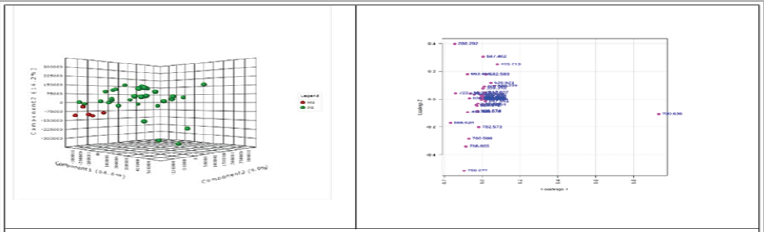
These graphics show a good discrimination between PS and MS groups (PC1=64.6%, PC2=14.2%, PC3= 9.9%) between both groups (Figure 1A). The most important molecules identified have m/z values of 700.636, 647.462, 635.504, 415.213, 614.574 and 579.537. From the multivariate classification through PLS- DA, the performance of PLS regression coefficient in selecting relevant predictors were analyzed a supervised class prediction model, in order to determine the capacity for global metabolomes to accurately classify BC patients into their respective groups and to identify the most important metabolites to the class prediction and having the strongest correlation to BC diagnosis and evolution. These unsupervised multivariate statistics display the overall data variance in the datasets (PCA), whereas supervised statistics (PLS- DA) tries to identify the molecules which contributed most to the discrimination between groups PS vs MS, M vs PM, as well between different cancer stages (I, II and III).
Figure 2 represents the metabolites which contribute significantly to group discrimination classified after PLS regression coefficient (left) and the hierarchical clustering analysis (heatmap) using the significant metabolites initially selected (right) related to MS and PS groups. According to Figure 2, significant modifications between PS and MS groups were noticed, with PLS regression coefficients higher than 40%. Quantitative decreases in PS vs MS group molecules were noticed, to mention m/z= 415.213 identified as octacosapentaenoic acid (28:5), for 16, 16-dimethyl-PGD2/E2 (m/z=381.300), PC (36:4) (m/z=782.573), PC(16:0/9:0(COOH)) (m/z=666.628) and PC(38:6) (m/z= 806.574). Meanwhile other molecules (from diacyl glycerol, PC and LPC classes) had increased peak area values, to mention m/z=614.574, 579.537, 551.506, 586.542, 496.341, 642.607 and 758.572, identified and presented in (Table 1).
Figure 2: Metabolites which contribute to PLS regression coefficient classification (left) and Hierarchhical clustering analysis using the statistical significant metabolites (right) related to MS and PS groups.
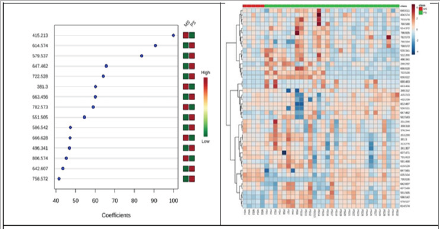
Table 1: Identification of lipid molecules which were selected by unsupervised multivariate analysis. The identification was based on Lipid maps and Human Metabolomic Data Base.
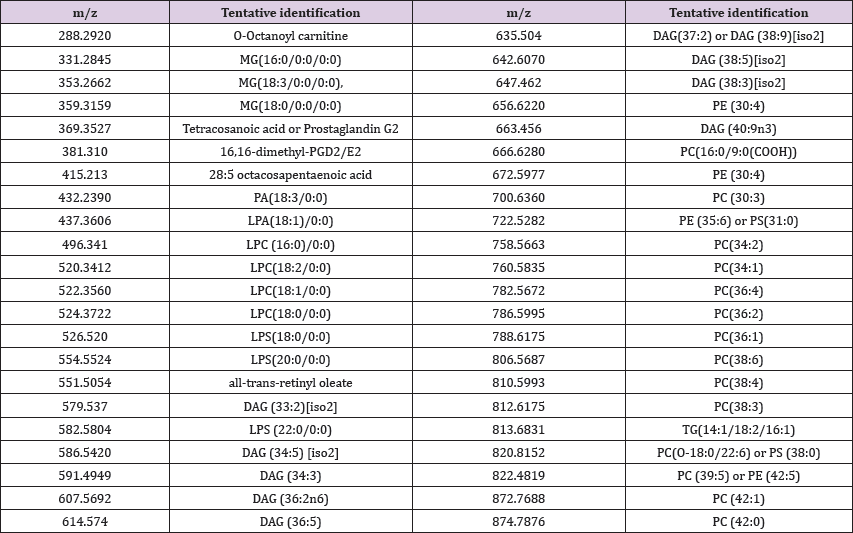
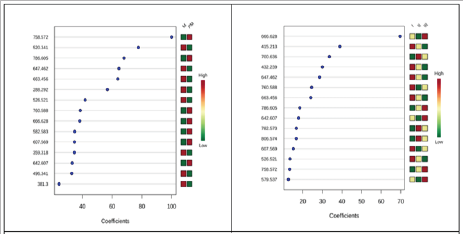
Table 1 presents the tentative identification of lipid molecules, initially selected by multivariate analysis. The identification was made by consulting the Lipid Maps and Human Metabolomics Data Bases (Table 1). (Figures 3A & 3B) represents the metabolites which contribute to PLS regression coefficients classification related to M and PM groups (left) and to different BC stages (I, II z=288.292), LPS(18:0/0:0) (m/z=526.521) MG(18:0) (m/z= 359.318), LPC (16:0)/0:0) (m/z= 496.342) and 16,16-dimethyl- PGD2/E2 (m/z=381.300).
Meanwhile, increases of of peak areas for PM group comparing with M groups for PC (34:2) (m/z=758.572) , PC(36:2) (m/z= 786.605), PC(34:1), ( m/z=, 760.588), PC(16:0/9:0(COOH)), (m/ z= 666.628) LPS(22:0/0:0) (m/z=582.583), DAG (36:2n6) (m/ z=607.569) and DAG (38:5)[iso2]. (m/z= 642.607). According to (Figure 3 B) significant number of compounds with PLS regression coefficients higher than 10, gradual increases from stage I to II and III were noticed: PC(36:2) (m/z= 786.605), and PC(34:2) (m/ z=758.572), while gradual decreases from I to II to III for m/z for PA(18:3/0:0) (m/z= 432.239), DAG (38:3)[iso2] (m/z=647.462 ), DAG (40:9n3) (m/z= 663.459) and LPS(18:0/0:0) (m/z=526.521). The evolution from stage I to II, was marked by increases of PC (30:3) (m/z= 700.736), PC(36:4), (m/z= 782.573) and PC(38:6) (m/z= 806.574), and decreases of PC(34:1) (m/z= 760.588) and DAG (36:2n6) (m/z=607.569). Between stages II and III here were identified mainly increases for PC(16:0/9:0(COOH)) (m/z = 666.628), DAG (38:5)[iso2](m/z=642.607), DAG (33:2)[iso2] (m/ z= 579.537).
Figure 3: Metabolites which contribute to PLS regression coefficient classification related to M and PM groups (left) and to different BC stages (I, II and III) (right).
The results obtained by multivariate analysis were further analyzed and complemented by fold change, T-test (evaluation of P-values and their significance) and the predictive value of some metabolites was performed by ROC analysis (sensibility-specificity matrix), calculating the areas under the ROC curve (AUROC values). Table 2 includes the mean fold change values molecules initially selected by Profile Analysis 2.1. For groups MS vs PS, M vs PM, in order to evaluate the tendency (decrease or increase) of such molecules between these groups (Table 2). Tables 3 & 4 includes also the data obtained by univariate analysis using Malabo analyst 3.0, showing the significant differences and tendency (decrease or increase) of selected metabolites based on their highest AUROC values, molecules only with P-values <0.05, Log2 fold change (FC) and highest PLS regression coefficients.
Table 2: The mean fold change (FC) values for molecules separated from groups PS/MS and M/PM, to evaluate the tendency (decrease or increases of more than 15% are bolded).

Table 3 refers to statistical differences between MS and PS groups, while (Table 4), the evolution between BC stages I, II and III and M vs PM (Tables 3 & 4). When compared groups PS and MS, seven molecules showed AUROC values above 0.819, the most predictive ones being considered PC(30:3) and five DAG derivatives of unsaturated fatty acids (37:2 or 38:9, 34:5, 36:5, 38:3, 33:2) with significant P values <0.02. When compared the molecules related to BC evolution from stage I to II, II to III, I to III, five molecules showed AUROC values between 0.759 and 0.732, the most predictive being PC( 16:0/9:0 COOH), and three DAG of unsaturated fatty acids (36:5, 38:3, 34:5) with significant P values <0.02. When compared groups M and PM, only one metabolite showed significant differences, LPC(18:2/0:0) with an AUROC value of 0.676 and P = 0.068.
Table 3: Molecules selected by Metaboanalyst 3.0: univariate analysis comparison between MS and PS groups: the area under ROC curve (AUROC), P value (T-statistics) and Log2 fold change (FC) for metabolites with PLS regression coefficients) higher than 40 and P values <0.05.
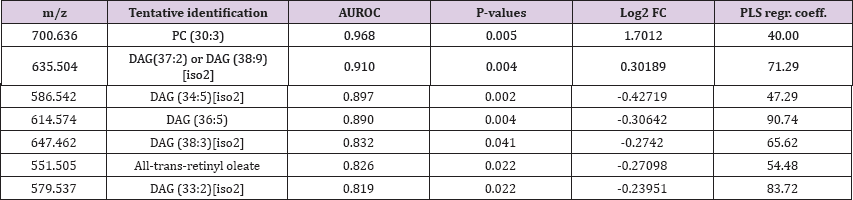
Table 4: Comparisons between the stages I to II, II to III, I to III and M to PM groups. The metabolites displayed are ranked based on area under ROC curve (AUROC), T-statistics and Log2 fold change (FC). The metabolites displayed in the table are ranked based on highest area under ROC curve (AUROC), T-statistics and Log2 fold change (FC).

Increased de novo lipid synthesis is typically found in tumor cells but not in normal cells, which would make it a promising target for new tumor-specific therapeutic approaches, including breast cancer. Higher levels of lipid metabolites, including fatty acids, phospholipids and their metabolites, have been found in BC patients compared to controls. Wang et al. [6] reported that several lipids including phosphatidyl glycerol (PG), Phosphatidylcholines (PCs) were upregulated in highly invasive BC cells, the tumors developing a 'lipogenic phenotype' with an upregulated lipogenesis and lipolysis, associated with tumor aggressiveness [9,25,26]. Specifically, PCs of saturated FAs (C14:0, C16:0 and C18:0) were found at higher levels in aggressive tumors, and predicted poorer survival for BC patients. Human plasma metabolomics was applied for identifying differential metabolites released from cells, as a mirror of the cell activity, useful to predict BC and its molecular subtypes [27].
It is well documented the upregulation of de novo fatty acid synthesis used to build membrane phospholipids, needed in rapidly proliferating BC cells and for cell signaling, therefore de novo FAs synthesis indirectly affect cell signaling processes. Meanwhile, phosphatidyl and eicosanoid lipids synthesis in BC cells is upregulated, supporting the cell proliferation by activation of Fatty acid synthetase (FAS) and protein kinase C (PKC), a is a stimulator of diacylglycerol formation [28,29]. Phosphatidylcholines (PCs) are known as an important cellular signaling molecules which may generate diacylglycerols by lipolysis in hormone- stimulated cells, by a phospholipase C- mediated mechanism [30]. Diacylglycerols (DAGs) including different fatty acids exhibit as well signaling functions [31]. In this context, our results reflect, by successive multivariate and univariate statistics that the potential lipid biomarkers responsible for the discrimination of serum composition between IDC-BC patients and healthy controls belong mainly to DAG and PC classes, as derivatives of different unsaturated fatty acids.
Further studies need to elucidate the importance of the balance between saturated and unsaturation of FAs which esterify DAG and PCs. The BC evolution from stage I to III may be predicted also by a PC molecule and three DAG derivatives, while discrimination between pre-menopause (M) and menopause (PM) patients can be predicted only by a potential biomarker, LPC (18:2/0:0). These preliminary results are complementary to previous findings showing the important role of phospholipid metabolic pathways in breast cancer diagnosis and evolution, as well the signaling role of diacylglycerols as intracellular lipolysis markers, identified in blood serum. It is obvious that such data need validation for larger cohorts of patients in order to establish a significant number of relevant biomarkers for breast cancer diagnosis and prognosis. It is obvious also that clinical application of lipidomics' approach can be developed and further improved by integration of lipidomics data with genomics and proteomics, thus developing new frontiers in biomarker discovery for breast cancer.


