Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Thongchai Taechowisan*1, Jakkrit Jantiya1, Nutthapol Mungchukeatsakul1, and Waya S Phutdhawong2
Received: March 9, 2018; Published: April 04, 2018
Corresponding author: Thongchai Taechowisan, Department of Microbiology, Faculty of Science, Silpakorn University, Nakorn Pathom 73000,Thailand
DOI: 10.26717/BJSTR.2018.03.000910
Sweet Basil (Ocimum basilicum Linn.) has a variety of therapeutic properties, including antimicrobial activity. This study aimed to determine the antibacterial activity of the major chemical components in the crude extract against methicillin resistant Staphylococcus aureus (MRSA) isolated from the clinical specimens. TLC and column chromatography resulted in the separation of two major compounds. On the basis of the spectral data, they were identified as linalool (1) and 1,8-cineole (2). Antibacterial activity of crude extract and isolated compounds was evalusted by disc diffusion, MIC and MBC against 9 multi-drug resistance S. aureus isolated from clinical specimens. The crude extract and isolated compounds exhibited an excellent antimicrobial activity at a dose level of 34.5 mg/disc for disc diffusion method. The crude extract and linalool showed higher activity against tested MRSA with MIC (<0.09 mg/ml) and MBC (≤ 0.09 to 0.38 mg/ml) than 1,8-cineole. The results suggest once again that the antimicrobial activity of the crude extract and major isolated compounds, linalool and 1,8-cineole are a resultant of the antibacterial property against MRSA.
Keywords: Antimicrobial Activity; Chemical Composition; Methicillin-Resistant Staphylococcus Aureus; Ocimum Basilicum L
Abbreviations: MRSA: Methicillin Resistant Staphylococcus Aureus; NMR: Nuclear Magnetic Resonance UV : Ultra Violet; TLC: Thin Layer Chromatography; BA: Blood Agar; MSA: Mannitol Salt Agar; MIC: Minimum Inhibitory Concentration; MBC: Minimum bactericidal concentration; PCR: Polymerase Chain Reaction
Now a day's multi-drug resistance has developed due to the uncritical use of commercial antimicrobial drugs commonly used in the treatment of infectious disease. In addition to this problem, antibiotics are sometimes associated with adverse effects on the host including hypersensitivity, immune-suppression and allergic reactions. This situation forced scientists to search for new antimicrobial substances. Given the alarming incidence of antibiotic resistance in bacteria of medical importance, there is a constant need for new and effective therapeutic agents. Therefore, there is a need to develop alternative antimicrobial drugs for the treatment of infectious diseases from medicinal plants Agarwal et al. [1]. Ocimum basilicum L. (sweet basil) belongs to the family Lamiaceae, distributed throughout the tropical and subtropical regions of Asia, Africa and Central and South America [2]. Secondary metabolites from Ocimum species possess exceptional biological activity and have antimicrobial [3-6] and antioxidant [7-9]; bactericidal [10,11] repellent [12-13], anticonvulsant [14], chemopreventive and radioprotective effects [15-16].
The chemical composition of essential oils of Ocimum species has been well studied. As prevalent components many basil essential oils contained monotherpene derivatives (camphor, limonene, 1,8-cineole, linalool, geraniol) and phenylpropanoid derivatives (eugenol, methyleugenol, chavicol, estragole, methyl- cinnamate) [4,17-19]. Different chemotypes of basil have been recognized based on the predominant essential oil constituents (e.g. linalool, methylchavicol, methyl cinnamate, methyleugenol, eugenol) [17,20]. There are only few data regarding the antimicrobial activitiy of essential oils of Ocimum species against methicillin-resistant Staphylococcus aureus (MRSA), especially MRSA from the clinical isolates. This paper presents the major chemical composition of the essential oils of Ocimum basillicum Linn and to evaluate the antibacterial effects on MRSA for providing a basic research data needs and for practical use.
By using a UV lamp and Iodine crystals. The results showed two major spots with Rf = 0.60 and 0.83. Preparative TLC on the oily material was carried out and the two fractions with Rf = 0.60 and 0.83 separated and characterized. IR, UV-Visible, 1H-NMR and MS spectra of each fraction was taken. Ethics statement. The study was approved by the Committee of the Scientific Study of Humane Technique in Laboratory Animal Experiments and Human Ethics, Faculty of Science, Silpakorn University, Nakhon Pathom, Thailand. No written informed consent was required because all the patients were anonymous and any other personal information was not used in this study. Plant material and extraction procedure. Leaves of sweet basil (Ocimum basillicum L.) were collected from Nakorn Pathom, Thailand. The dried leaves (500 g) were extracted successively with 1 L of ethyl acetate for overnight at 30oC.
The aqueous extracts were filtered using Whatman filter paper (No.1) and then concentrated in vacuo at 40oC using a Rotary evaporator. The residues obtained were stored in a freezer at -20oC until further studies. Isolation of the chemical components of the crude extracts. A glass column 48 inches in height and 1.0 inch internal diameter was used for the chromatography of the crude extracts. About 5 g of the crude extract was chromatographed in a column using 40 g of silica gel (60-120 mesh) as stationary phase. The extract was eluted with increasing polarity of ethyl acetate and chloroform mixtures successively using them as elements (10:0, 9:1, 8:2, 7:3 up to 0:10) to choose the appropriate solvent for the complete resolution of the spots. Thirty millilitre fractions were collected. All fractions were monitored by thin layer chromatography (TLC). Fractions with spots of the same retention factor (Rf) values were combined and rechromatographed in appropriate solvent systems until pure isolates were obtained.
Spots on the TLC plate were visualized using Ultra Violet (UV) lamp (254 nm and 365 nm) with some amount of iodine vapor. A glass column 24 inches in height and 0.5 inch internal diameter was used for the rechromatography of smaller fractions from the first column. Ten millilitre fractions were collected. Final purifications were conducted using 20 millilitre pipettes as columns. Two millilitre fractions were collected. Finally, IR and one dimentional Nuclear Magnetic Resonance (1H-NMR, 13C-NMR) and DEPT spectroscopic techniques were used to characterize the isolated compounds. TLC analysis on silica gel with chloroform: ethyl acetate (5:1) as the mobile phase was carried out.
The spots were developed Characterization of the component with Rf = 0.60 : IR (vmax, cm-1, neat liquid): 3450 s (OH); 2968 s, 2922 s (C-H, aliphatic), 1637 m-w (C=C), 1450 (CH2), 1374 (CH3); UV CH2Cl2 λmax = 227 and 220 nm; 1H-NMR (CDCl3, 500 MHz) δ(ppm): 1.27 [CH3(C-3), s], 1.57 [CH2(C-4), m], 1.60 [CH3(C-7), s], 1.67 [CH3(C-7), s], 1.92 [OH(C-3), s], 2.04 [CH2(C-5), m], 5.05 [H(C-1), d], 5.11 [H(C-6), t], 5.21 [H(C-1), d], 5.91 [H(C- 2), dd]; 13C-NMR (CDCl3, 125 MHz) δ(ppm): 17.692 (C5), 22.825 [CH3(C-7)], 25.705 (C8), 27.812 [CH3(C-3)], 42.116 (C4), 73.447 (C3), 111.690 (C1), 124.407 (C6), 131.822 (C7), 145.094 (C23); MS (EI) showed m/z: 137 [(M-OH)+ , 2.1 %], 136 [(M-H2O)+ , 4.3 %], 121{[M-(CH3+H2O)]+ , 15.1 %}, 83{[CH2=CH-C(OH)(CH3)CH2] + , 12.2 %}, 71 {[(CH3)2C=CH-CHCH2] + , 82.2 %}, 69 {[CH2=CH-C(OH) (CH3)]+ , 38.8 %}, 55 {[(CH3)2C=CH]+ , 60.4 %}, 51 {[69-H2O]+ , 5 %}, 43 {[CH2=CH-C(CH3)]+ , 100 %}. On the basis of these results, this component was identified as linalool (1).
Characterization of the component with Rf = 0.83 : Its IR (vmax, cm-1, neat liquid): 2966 s, 2942 s, 2922 s (C-H, aliphatic), 1464 s, 1445 s (CH2), 1374 s, 1359 s (CH3), 1214 s, 1166 s (C-O); UV CH2Cl2 λmax = 227 and 212 nm; 1H-NMR (CDCl3, 500 MHz) δ(ppm): 1.048 [CH3(C-1), s], 1.24 [2 x CH2(C-3), s], 1.41 [H(C-4), s], 1.53 [4H(C- 7, C-8), q], 1.66 [2H(C-5/C-4), t], 2.04 [2H(C-4/C-5), t]; 13C-NMR (CDCl3, 125 MHz) δ(ppm): 22.85 [2 * CH3 (C3)], 27.59 [CH3(C-1)], 28.90 [(C7 + C8)/(C5 + C6)], 31.52 [C5 + C6)/(C7 + C8)], 32.94 (C4), 69.75 (C3-O), 73.59 (C1-O); MS (EI) showed m/z: 154 [M+ , 89 %], 140 [(M-CH2) + , 7.8 %], 134 [(M-CH3) + , 83.5 %], 126 {[140- (CH2)]+ , 10.7 %}, 111 [98 %], 96 [52.8]+ , 68 [45.5 %]+ , 67 [29.2 %]+ , 58 {[(CH3)2C=O]+ , 29.2 %}, 43 {[CH3CO]+ , 100 %}. On the basis of these results, this component was identified as 1,8-cineole (2). Bacterial isolates. Methicillin-resistant Staphylococcus aureus (MRSA) was isolated from clinical specimens (blood, sputum, pus, wound, urine and nasopharyngeal swab samples) of patients admitted to Nakorn Pathom Hospital, as described in our previous report [21].
Briefly, isolates were examined by conventional methods such as colony morphology on blood agar (BA) and mannitol salt agar (MSA), Gram stain characteristics, catalase production and mannitol utilization then identified as S. aureus by tube coagulase test. Identified strains were stored at -20oC in Nutrient broth (Oxoid) containing 20% glycerol. Determination of methicillin resistance. Methicillin resistance was evaluated using three methods: 1) Disk diffusion test using 30 μ g cefoxitin disk (≤ 21 mm indicated MRSA); 2) Oxacillin MIC (Minimum Inhibitory Concentration) test (≥ 4 μ g/ml indicated MRSA); and 3) Polymerase chain reaction (PCR) for the detection of mecA gene (positive indicated MRSA) [22]. Antibiotic disks and Oxacillin powder were obtained from Himedia (Himedia Laboratories, Pvt. Ltd., Mumbai, India). All tests were compared for sensitivity and specificity with PCR for mecA gene as reference method. Sensitivity was calculated by dividing the number of mecA-positive isolates detected as resistant using phenotypic methods by the total number of mecA-positive strain (ether susceptible or resistant). Specificity was calculated through dividing the number of mecA-negative isolates classified as sensitive based on phenotypic criteria by the total number of mecA-negative isolates [23].
Antibiotic Sensitivity test, Antibiotic susceptibility of the bacteria isolates was assayed according to the Kirby - Bauer disk diffusion method [24]. All the plates were incubated for 20 minutes before inoculation and placement of antibiotic disc to allow excess moisture to dry. After the drying, a single loop of each isolate was inoculated into sterile normal saline and compare with 0.5 McFarland standard, the suspension was aseptically swabbed on the surface of Mueller-Hinton plates and antibiotic sensitivity disk that contain penicillin (10 unit), cefoxitin (30 μ g), chloramphenical (30 H g), tetracycline (30 μ g), erythromycin (15 μ g), trimethoprim/ sulfamethoxazole (25 ng), gentamicin (10 ng), clindamycin (2 μ g), rifampicin (30 μ g), linezolid (30 μ g) and ciprofloxacin (25 μg). MICs of oxacillin and vancomycin to the both MRSA and MSSA isolates were determined by broth dilution method.
All procedures were carried out and interpreted according Clinical and Laboratory Standards Institute guideline CLSI. S. aureus ATCC25923 was used as control strain in disk diffusion and broth dilution methods. PCR amplification. Total bacterial DNA was extracted from S. aureus using a modified phenol - chloroform method of Tiago. Briefly, S. aureus was cutured on 5 ml brain heart infusion broth and incubated at 37oC for 24 hours. Afterwards,1.5 ml of the culture was centrifuged for 5 minutes at a velocity of 14,000 rpm. Pellet was resuspended in 600 μ l Tris-EDTA buffers by repeated pipetting or overtaxing, then 3 μ l of 10% SDS and 3 H l of 20 mg/ml proteinase K were added, mixed and incubated for 30 minutes at 65oC in water bath. A volume of phenol/chloroform/ isoamyl alcohol equal to (600 μ l) was added, mixed, centrifuged for 5 minutes at speeding (14,000 rpm) and the supernatant was transferred to fresh tube, then equal volume of ethanol was added and mixed gently until DNA precipitated, then centrifuged for 5 minutes and the supernatant was discarded.
The pellet was washed with 1 ml of 70% ethanol, mixed, centrifuged for 5 minutes, and the supernatant was discarded, then it was dried for 10 minutes at a velocity vac/45oC. Finally it was resuspended in 30 μ l TE buffer. DNA concentration was read using 2 n l in Nanodrop machine using TE buffer as blank and the concentration was made up to 100 ng/ μ l in each sample and was stored at -20oC. Primers used for detection of the femA gene were primers FemA1 and FemA2, leading to an S. aureus-specific 450- bp PCR product [25], while the mecA gene was detected with the primers MecA1 and MecA2, yielding a 519-bp PCR product for methicillin and oxacillin resistance [26] the aac(6’)/aph(2”) gene was detected with the primers aac(6’)/aph(2”)1 and aac(6')/ aph(2”)2, yielding a 407-bp PCR product for gentamicin resistance [27], the blaze gene was detected with the primers blaZ1 and blaZ2, yielding a 774-bp PCR product for penicillin resistance [26], the ermA gene was detected with the primers ermA1 and ermA2, yielding a 190-bp PCR product for erythromycin resistance, and the tetK and tetM genes were detected with the primers tetK1 and tetK2, yielding a 360-bp PCR product and primers tetM1 and tetM2, yielding a 158-bp PCR product, respectively [28].
The following reaction mixture was added to each sample: 2 μ l DNA (100 ng), 2 μ l primer (100 pmol), PCR mixture (1.5 μl MgSO4,2.5μ l 10xPCR buffer, 0.5 μ l dNTPs, 0.2 μ l Taq polymerase) and completed to 25 μ l volume by H2O. The primer sequences and the PCR conditions were showed in Table 1. For the visualization of the product, 10 μ l of the each PCR reaction was mixed with 5 μ l 6x loading dye and loaded on 1.5% agarose gel for electrophoresis and visualization of the amplified PCR products. A 100 bp molecular weight DNA ladder was used for the validation of length of the amplified products (Vivantis Technologies) 002E Antibacterial assay. The tested of methicillin-resistant Staphylococcus aureus isolates were ptocessed for susceptibility tests using the method of Bauer-Kirby. Sterile paper discs (6 mm, Whatman 2017-006) were loaded with 50 μl of two-fold dilution of 275.6 mg/ml of each of the tested crude extract and purified compounds. Dilutions of bacterial suspensions were prepared using McFarland standard tubes (1 x 108 CFU/ml).
Table 1: Primer sequences and PCR conditions used.
Sterilized plates containing Muller Hinton agar were loaded with the bacterial suspension and the discs were loaded with various concentration of the tested crude extract and purified compounds. The loaded discs were aseptically placed on the inoculated agar plate's surface. All plates were incubated at 37oC for 24 h then the diameter of the formed inhibition zones around each disc was recorded and compared according to Clinical Laboratory Standard Institute (CLSI, 2012) [29]. The experiment was carried out in triplicates using controls. Minimum inhibitory concentration (MIC) and Minimum bactericidal concentration (MBC) determinations. The MIC was determined by micro-broth dilution methods [30]. The tested extract and purified compounds were serially diluted in nutrient broth containing 0.05% phenol red and supplemented with 10% glucose (NBGP) medium in 96-well plates.
Duplicate wells of each dilution (0.09, 0.18, 0.38, 0.73, 1.47, 2.94, 5.88, 11.75, 23.5, 47 and 94 mg/ml) were inoculated with 5 x 105 cells of the tested MRSA strain then cultures were incubated at 37oC for 18 h. Microbial growth was determined by observing the change of colour in the wells (red when there is no growth and yellow when there is growth). The lowest concentration showing no colour change was considered as the MIC. For the determination of MBC, a portion of liquid (10 μl) from each well that showed no change in colour was plated on MHA and incubated at 37oC for 24 h. The lowest concentration that yielded no growth after this sub- culturing was taken as the MBC.
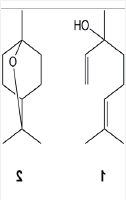
TLC and column chromatography on silica gel with chloroform: ethyl acetate as the mobile phase resulted in the separation of two major compounds with Rf = 0.60 and Rf = 0.83. 1H-NMR, 13C-NMR, UV-visible, IR and MS spectra of these compounds were taken. On the basis of their spectra, the compound with Rf = 0.60 was identified as linalool (1) and the compound with Rf = 0.83 was identified as 1,8-cineole (2). The structures of these compounds are shown in Figure 1.
Figure 1: The structures of these compounds
The tested 9 methicillin resistant S. aureus isolates has shown to be multi-drug resistant. The multi-drug resistance ranged from 3 to 6 antibiotics (Table 2). Two MRSA strains were resistant to 6 antibiotics, 3 were resistant to 5 antibiotics, were resistant to 4 antibiotics, and only one strain was resistant to 3 antibiotics.
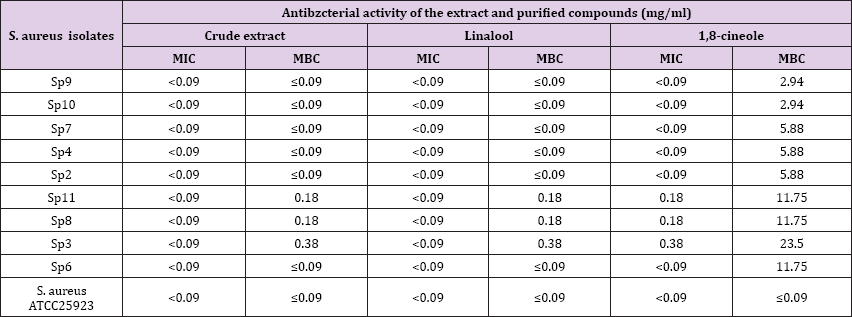
The susceptibility of the crude extract and purified compounds showed that the inhibition zone diameter ranged from 10.3 to 22.3 mm. The highest effect was exerted by linalool (22.3 ± 0.23 mm) on MRSA strain Sp9 which was isolated from sputum specimen, the least effect was recorded by crude extract (10.3 ± 0.64 mm) on MRSA strain Sp3 which was isolated from pus specimen (Table 2). The minimum inhibitory concentration (MIC) values of the crude extract and purified compounds ranged between < 0.09 to 0.38 mg/ml. The most potent inhibition was recorded to the crude extract and linaloool with < 0.09 mg/ml, while the least potent inhibition was 1,8-cineole with 0.38 mg/ml on MRSA strain Sp3. The minimum bactericidal concentration (MBC) values of the crude extract and purified compounds ranged between ≤ 0.09 to 23.5 mg/ ml. The most potent MBC was recorded to the crude extract and linalool with ranging between ≤ 0.09 - 0.38 mg/ml, while the least potent MBC was 1,8-cineole with 23.5 mg/ml on MRSA strain Sp3 (Table 3).
Table 2: Correlation between susceptibility pattern (inhibition zone diameter) of crude extract, purified compounds and antibiotic resistant pattern and PCR results of methicillin-resistant Staphylococcus aureus isolates.

aPEN, penicillin; ERY, erythromycin; TE, tetracycline, FOX, cefoxitin; GEN, gentamicin; CIP, ciprofloxacin; SXT, trimethoprim- sulfamethoxazole; OXA, oxacillin; DA, clindamycin.
Table 3: Minimum inhibitory concentration and minimum bactericidal concentration values of crude extract, purified compounds on methicillin-resistant Staphylococcus aureus isolates
The increasing incidence, of S. aureus antibiotic resistance particularly in hospitals towards methicillin and wide range of other antimicrobial agents, resulted in announced therapy difficulty [31]. Methicillin-resistant Staphylococcus aureus (MRSA) constitute a human health problem causing a real life threat to hospitalized patients. Hospital acquired S. aureus infection still difficult to treat with known antibacterial agents consequently, several attempts were exerted to control this phenomenon spread through the search for the discovery of alternative effective herbal therapy for treating the infection with MRSA [30]. Medicinal plants became targeted for the production of new effective antibacterial drugs production for overcoming the extensive occurrence of multi-drug resistance. Sweet basil (Ocimum basilicum L. plant of the family Lamiaceae) is an aromatic, annual herb. Traditionally, it is used for medicinal purposes as a digestive stimulant and for treatment of headaches, coughs, diarrhea, insomna, constipation, warts, wounds, and kidney malfunction [32,33].
Basil leaf oil is used principally in the food and cosmetic industries [33]. It has wide applications as a spice in a variety of foods, beverage, and confectionary products. It also possesses antimicrobial activity and biologically active [34,35]. It has been postulated previously that this herbal extract might work through inhibition of different cellular biotic functions including ion leakage from the cells and disrupting the plasma membrane permeability [36]. The major components of basil oil vary extensively, depending on genetic factors, cultivars, geographical origins, nutritional status, the extracted plant materials (stem, leaf, and flower), extraction methods, and so on [3,37,38]. Therefore, the present study was conducted to analyze the major composition of leaves extract of O. basilicum L. and to evaluate its antimicrobial activity against MRSA from the clinical isolates.
There are many varieties of O. basilicum, as well as several related species or species hybrids also called basil. The type used in Italian food is typically called sweet basil, as opposed to Thai basil (O. basilicum var. thyrsiflora), lemon basil (O. citriodorum) and holy basil (O. tenuiflorum), which are used in Asia. While most common varieties of basil are treated as annuals, some are perennial in warm, tropical climates, including holy basil and a cultivar known as African Blue. Basil is originally native to India and other tropical regions of Asia, having been cultivated there for more than 5,000 years [39]. The basil comes in many different varieties, each with its own unique chemical composition and characteristic flavor. The main use of the herb is culinary. The flavour and character of any particular variety of basil is affected to a great extent by many external environmental factors, including factors such as temperature, the type of soil, the geographic location, and even the amount of rainfall received by the individual plant.
Basil extract has various chemical compounds that include α-pinene, camphene, β-pinene, myrcene, limonene, cis-ocimene, camphor, linalool, methyl chavicol, γ -terpineol, citronellol, geraniol, methyl cinnamate and eugenol and other terpenes [40]. Methyl eugenol [1,2-dimethoxy-4-(2-propenyl) benzene, found in sweet basil is a member of a family of chemicals known as ally alkoxy-benzenes, which include other naturally occurring materials such as isoeugenol, eugenol, estragole, and safrole. All these compounds typically enter the diet via a variety of different food sources, including spices (nutmeg, all spices), herbs (basil, tarragon), bananas and oranges. Many of these compounds are also found as components of natural oils used in perfume [41-43]. 3,7-dimethyl-1,5-octadien-3,7-diol is a volatile secondary alcohol found in wine and produced during fermentation and was also reported as a constituent of sweet basil [44].
Two isomeric compounds named (1-(2-vinylcyclohexa-1,4- dienyl) propan-2-ol and 2-(2-vinylcyclohexa-1,5-dienyl) propan- 1-ol were isolated from sweet basil leaf extract [45]. In this study, the major chemical composition, linalool and 1,8-cineol were isolated from the leaves extract of O. basilicum L. The isolation of linalool and 1,8-cineol from the crude extract of O. basilicum has been previously reported [46-48] whilst other reports were also isolated from Ocimum canum, Ocimum gratissimum [38], Ocimum kilimandscharicum, Ocimum tenuiflorum [46,49] and Salvia officinalis L. (family Lamiaceae) and TTachyspermum ammi L. (another plant of the family Apiaceae) [50], Thymus mastichina L., T. camphoratus and T. lotocephalus (family Lamiaceae) [51] and Lavandula angustifolia Miller and Lavandula x intermedia (family Lamiaceae) [52]. Basil essential oil is traditionally classified into four distinct chemotypes with many subtypes, based on the biosynthetic pathways that produce the principal components in the oil.
The different chemotypes contain various proportions of allyl- phenol derivatives, including estragole (methylchavicol), eugenol, and methyl eugenol, as well as linalool, a monoterpene alcohol [20,53,54]. The antimicrobial activity of the individual principle constituents of basil essential oil (linalool, methyl chavicol, eugenol, and methyl cinnamate) was also studied. Prasad et al. [35] studied the antimicrobial activity of essential oils of O. basilicum (French), O. basilicum (Indian), and O. basilicum (Niazbo), which are rich in linalool, methyl chavicol, and methyl cinnamate, against 11 Gram positive and 7 Gram negative bacteria. They found that these oils were more effective against Gram positive bacteria, Bacillus sacharolyticus, B. stearothermophilus, B. subtilis, B. thurengiensis, Micrococcus glutamicus, and Sarcina lutea were inhibited by each of these basil essential oils. Studies in the literature suggest that linalool, a monoterpene, is the main ingredient responsible for antibacterial activity [55]. Knobloch et al. [56] evaluated the antimicrobial activity of essential oil components against Gram negative bacteria (e.g. Enterobacter aerogenes and Proteus vulgaris), Gram positive bacteria (e.g. S. aureus and Bacillus subtilis), and fungi (e.g. Aspegillus flavus, A. Niger, A. ochraceus, and Penicillium expansum).
They found that linalool, with its high water solubility, had a significant antimicrobial activity as compared to cinnamaldehyde, citral, geraniol, eugenol, and menthol whereas methyl chavicol, with its lower water solubility, had a low antimicrobial activity. The solubility in water of essential oil constituents is directly related to their ability to penetrate the cell walls of a bacterium or fungus. Thus, the antimicrobial activity of essential oils is due to their solubility in the phospholipids bilayer of cell membranes [56]. It has also been suggested that linalool has the potential to act as either a protein denaturing agent or as a solvent dehydrating agent who may also contribute to its antimicrobial activity [57]. It was also reported that the antibacterial activities of monoterpene alcohols (including linalool, nerol, citronellol, and geraniol) are more effective than their antifungal activity.
Meena and Sethi [58] found that eugenol has an inhibitory effect against A. Niger, Lactobacillus acidophilus, and Saccharomyces cerevisiae. Kim et al. [59] studied the antibacterial activity of some essential oil components (including linalool and eugenol) against food-borne pathogens (E. coli, E. coli O157: H7, Salmonella typhimurium, Listeria monocytogenes, and Vibrio vulnificus). They found that eugenol showed a dose-related increase in the zone of inhibition against the tested strains, whereas linalool exhibited a similar effect against all tested strains except for L. monocytogenes. Linalool inhibited the growth of L. monocytogenes, but the difference in the zone size between the test concentrations was not significant [59,60] studied the antibacterial properties of the aromatic constituents of essential oils. The results of the disk diffusion assays showed that linalool was the most effective compound and retarded 17 out of 18 bacterial strains (only VR- 6, a Pseudomonas sp., is resistant), followed by cineole, geraniol, menthol, and citral. They also found that the MIC values of the essential oils were usually lower than those of their constituents, as the similar finding in our study that the MIC values of the crude extract was lower than 1,8-cineole, but equal to linalool.
One possible reason for this result could be the synergistic action of the constituents in the crude extract. In studies by Bassole et al. [61], O. basilicum had as its main compounds: linalool and eugenol. The essential oil showed antibacterial activity against strains of S. aureus, E. faecalis, L. monocytogenes, E. aerogenes, E. coli, S. enteric, and S. typhimurium. Using the checkerboard method, associations of O. basilicum and eugenol showed synergetic effects, confirming the roles of certain components in the interaction. Based on the hypothesis of [62] we suggest that synergetic effects might be due to increases in one of three factors which determine monoterpene antimicrobial character: their lipophilic properties, the potency of their functional groups, and the resulting aqueous solubility when in a paired combination [56].
Mazzanti et al. [63] found that linalool was the active compound that completely inhibited the growth of tested yeasts (Candida albicans, C. krusei, and C. tropicalis), S. aureus and E. coli. Authentic pure linalool showed a similar antibacterial spectrum to that of basil essential oils. Dorman and Deans [64] reported on the antibacterial activity of 21 plant volatile oil components (including eugenol and linalool) against 25 bacterial strains by the agar well diffusion technique. Eugenol exhibited the widest spectrum of activity against 24 out of 25 bacteria, except for Lactococcus lactis subsp. cremoris, followed by linalool (against 23 strains, except L. cremoris and P aeruginosa). These results contradict those obtained by Lachowicz et al. [65] and Wan et al. [66] who used the same technique and found that linalool inhibited L. cremoris components with phenolic structures, including eugenol, were highly active against the test microorganisms.
Members of this class are known as either bactericidal or bacteriostatic agents, depending on the concentration [57]. These components are strongly active despite their relatively low solubility in water [56,59,67]. Alcohols are known to possess bactericidal rather than bacteriostatic activity against vegetative cells. The tertiary alcohol, linalool, is active against the test microorganisms, potentially acting as either a protein denaturating agent [57] or as a solvent dehydrating agent [68]. examined the antibacterial properties of the linalool and the methyl cinnamate type of the essential oils of O. basilicum, in the pure state and at ten-fold dilutions (1:10, to 1:10000) prepared with DMSO against four major bacterial species. They found that both essential oils had an inhibitory activity against S. aureus and E. coli at all dilutions. Basil essential oil also showed an inhibition against B. cereus, L. acidophilus, A. niger, and S. cerevisiae, as determined by the paper disk agar diffusion method, both at ambient temperature and 37oC [58].
These results were expanded and supported by Aboul Ela et al. [69]and Elgayyar et al. [70]who showed that basil essential oil has antibacterial and antifungal activity against S. aureus, E. coli, and A. niger. Our studies suggest the potential use of crude extract of Ocimum basilicum L., as antimicrobial substances for protection of the MRSA infection, food preservation and in the pharmaceutical industries [71]. However, further evaluation is needed in order to determine if these substances have a negative impact on normal human microbiota and also to investigate toxicities, and delineate mechanisms of action.
Sweet basil (Ocimum basilicum L.) crude extract and its major constituents, linalool and 1,8-cineole have a potential in antimicrobial activity against MRSA isolated from the clinical specimens. Especially, a higher antimicrobial effects of the crude extract and linalool compared with 1,8-cineole on MRSA.
This work was supported by Department of Microbiology, Faculty of Science, and Silpakorn University, Thailand. We gratefully acknowledge Department of Pathology, Nakorn Pathom Hospital, Nakorn Pathom, Thailand for providing clinical and environmental specimens for this work.


