Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Yahia A Kaabi*
Received: January 30, 2018; Published: February 08, 2018
Corresponding author: Yahia A Kaabi, Department of Medical Laboratory Technology, Jazan University, Saudi Arabia
DOI: 10.26717/BJSTR.2018.02.000741
Objectives: To establish whether 45-bp insertion (I)/deletion (D) polymorphisms in Uncoupling protein 2 (UCP2) are associated with moderate and/or severe obesity in a Saudi Arabian population.
Methods: The study enrolled 151 (male and female) subjects originating from the eastern province of Saudi Arabia, and assigned each to either a 'non-obese', 'moderately obese,' or 'severely obese' group. Genomic DNA was extracted from all subjects, and screened for UCP2 insertion/deletion polymorphisms using a standard PCR protocol.
Results: The overall frequencies of the UCP2 45-bp insertion/deletion genotypes D/D, I/D, and I/I within the analyzed population were 58.3%, 36.4%, and 5.3% respectively. The D/D genotype was highly prevalent within the severely obese group (82.9 %) compared to the non-obese (46.2%) and moderately obese (53.3%) groups. Using a dominance model, the conducted logistic regression analysis showed no significant association between the insertion allele and moderate obesity (OR = 0.75, 95% CI: 0.35-1.59, P = 0.585); however, in contrast, a strong association was found between the deletion allele and severe obesity (OR= 0.18, 95% CI: 0.07-0.44, P = 0.0004).
Conclusion: The present study reported the frequency of the UCP2 45-bp insertion/deletion polymorphisms in a population originating from eastern Saudi Arabia, and furthermore, identified a strong association between the D/D genotype and severe obesity in this population.
Keywords:Uncoupling protein 2 insertion/deletion polymorphism; Obesity
Abbreviations: UCP2: Uncoupling Protein 2; UTR: Untranslated Region; BMI: Body Mass Index; KFHU: King Fahad Hospital of the University; PCR: Polymerase Chain Response; SD: Standard Deviation; ORs: Odds Ratios
Obesity is a serious and increasing prevalent medical condition in both Western and some developing countries. It is associated with a high mortality rate, caused by complications including cardiovascular and inflammatory diseases, insulin resistance, and cancer [1]. For many Gulf countries, including Saudi Arabia, obesity is becoming a major health concern for patients of both sexes [2]. In fact, this condition is estimated to affect 24.1% of men and 33.5% of women in Saudi Arabia [3]. While risk factors such as a high caloric intake and a sedentary lifestyle are known to contribute to obesity pathogenesis, it is likely that predisposing genetic factors also contribute to the development of this condition. The mitochondrial proton carrier Uncoupling protein 2 (UCP2) is a member of the mitochondrial anion carrier protein subfamily, (which also includes UCP1, 3, 4, and 5). Uncoupling proteins function to increase proton influx through the inner mitochondrial membrane without ATP synthesis, and thereby enable efficient caloric consumption and heat generation [4].
They also inhibit the production of cellular free radicals and reactive oxygen species [5]. UCP2 is the most ubiquitously expressed of these proteins, and also exerts the greatest impact on mitochondrial function; thus, it has been the most extensively studied to date. The 8 kb UCP2 gene sequence is located on the q arm of chromosome 11, and comprises eight exons, and a GC rich promoter region [6]. Three polymorphisms have been previously identified in UCP2, including i) a -866 G/A polymorphism in the promoter region [7], ii) an Ala55Val polymorphism in exon 4, and iii) a 45-bp insertion/deletion polymorphism in the 3' untranslated region (3' UTR) of exon 8 [8]. Identified 45 bp insertion/deletion polymorphisms have not been shown to impact UCP2 mRNA expression; however, they do affect UCP2 mRNA stability [8,9], and are therefore likely to also impact UCP2 function, disrupting the normal rates of metabolism and fat deposition. Initially, the insertion allele was found to be associated with an increased metabolic rate and low body mass index (BMI) in a population of Pima Indians; however, several association studies have since been conducted to evaluate whether this insertion/deletion polymorphism is associated with obesity, and/or related disorders such as type II diabetes mellitus.
For example, two meta-analysis studies published in 2014 assessed the pooled effect of the UCP2 insertion/deletion polymorphism on the rate of obesity. The first of these found no significant association between the presence of the polymorphism and obesity in either European or Asian subjects [10], while conversely, the second revealed a significant association between the insertion allele and an increased body mass index (BMI) in analyzed Asian, but not European subjects [11]. It is likely that geographical differences continue to influence the pattern of distribution of UCP2 insertion/deletion polymorphisms, as well as their relatedness to obesity. This is supported by the fact that a recent study of an Iranian population (adjacent to Saudi Arabia), found a significant association between the homozygous deletion (D/D) genotype and metabolic syndrome [12]. Thus, the current study investigated whether the UCP2 45-bp insertion/deletion polymorphisms are associated with moderate and/or severe obesity in a Saudi Arabian population.
An informed consent was provided by all subjects for their participation in the study, whose design was approved by the institutional ethics committee (IRB-2015-03-167).
The present case-control study recruited 151 volunteer (male and female, aged 35-60 years) subjects that originated from the eastern province of Saudi Arabia, and attended the out-patient blood collection service provided by the King Fahad Hospital of the University (KFHU), at the University of Dammam, between February-July 2016. The subjects were divided into three groups, comprising
a) 'Non-obese' (BMI < 30 kg/m2, n=65),
b) 'Moderately obese' (BMI 30-35 kg/m2, n=45), and
c) 'Severely obese' (BMI ≥35 kg/m2, n=41).
Sociodemographic and health data for each subject (including gender, age, exhibition of type II diabetes mellitus and/or hypertension, and cigarette use), were provided via a structured questionnaire. Anthropometric data (including subject weight, height, and waist circumference) were measured using a metric and a vertical weight scale, with bare feet and only light clothing.
Each subjects BMI value was indirectly calculated according to the formula below:
BMI = Subject’s weight (kg) / (Subject's height (m))2
Whole blood samples (5 ml) were collected in EDTA vacationer tubes via aseptic venipuncture. Genomic DNA was then extracted from the leukocytes contained in 300 μL of each of these samples using an illustra blood genomic Prep Mini Spin Kit (GE Healthcare Life Sciences Ltd, UK) according to the manufacturer’s instructions.
The presence/absence of the UCP2 45-bp insertion/deletion polymorphism was assessed via polymerase chain response (PCR) using a 2x master mix (Promega; Wisconsin, USA), 1 μL of extracted template DNA, and 1 μL (0.1 ng) each of forward (5' CAG TGA GGG AAG TGG GAG G 3') and reverse (5' GGG GCA GGA CGA AGA TTC 3') primer. DNA amplification was achieved using a thermal profile comprising (initial denaturation) 4 min at 95°C, followed by 35 cycles of (denaturation) 30 s at 93°C, (primer binding) 30 s at 58°C, and (elongation) 30 s at 72°C, and a final (extension) step of 10 min at 72°C. The resultant products (10 μL) were then separated on a 1% agarose gel, and visualized using Ethidium bromide (EtBr) (0.5 μL/gel). Deletion (D) and insertion (I) alleles were identified to be present by the appearance of 412 bp and 457 bp bands, respectively (Figure 1).
Figure 1: The presence/absence of the UCP2 45-bp insertion/deletion polymorphism was confirmed by separating PCR products on a 1% agarose gel. Lane 1, DNA ladder (M); Lane 2 and 4, the homozygous deletion (D/D) genotype was detected as a single 412 bp PCR fragment; Lane 3, the homozygous insertion genotype (I/I) was detected as a single 457 bp PCR fragment; Lane 5 and 6, the heterozygous insertion/deletion (I/D) genotype was detected as two coincident 457 and 412 bp fragments.

All statistical analyses were performed using Graph Pad Prism 7 software (California, USA). Data from all study groups were presented as either the mean ± standard deviation (SD), or as percentage values. Continuous variables were assessed using an analysis of variance (ANOVA) test, whereas categorical variables were compared using a either a Chi-square or Fisher's exact test. Potential associations between the UCP2 insertion/deletion polymorphisms and obesity were evaluated by calculating the odds ratios (ORs), 95% confidence intervals, and two-sided Chi-square test and/or Yates' correction P values to conduct logistic regression analyses. A logistic binary regression was used to adjust all ORs to confound parameters such as subject age and sex. A P value less than 0.05 was considered to indicate statistical significance.
The demographic characteristics of the study population are summarized in Table 1. The mean age of the study population was found to be 44.78 ± 10.31 years, and no statistical difference in age was observed between the three study groups (P = 0.1023). The overall male/female ratio, which was 72/79 across the three study groups, was reduced in both the moderately and the severely obese subject groups compared to the non-obese group; however, these Table 1: Baseline characteristic of the study population, (n=151).differences were not statistically significant (P = 0. 1663). The mean (± standard deviation) BMI values for the non-, moderately, and severely obese subject groups were calculated to be 26.18 ± 1.92, 32 ± 1.83, and 39.59 ± 3.48 respectively, and shown to be significantly different via the conducted ANOVA test (P= <0.0001). Type II diabetes mellitus and hypertension were more prevalent in the moderately (affecting 40% and 28.9% of subjects, respectively) and the severely obese (affecting 51.1% and 53.65% of subjects, respectively) groups than in the non-obese group (affecting 29.2% and 18.5% of subjects, respectively). In contrast, the frequency of cigarette use was increased in the non-obese group (27.7%) compared to the moderately (13.3%) and severely obese (12.2%) groups.
Table 1: Baseline characteristic of the study population, (n=151).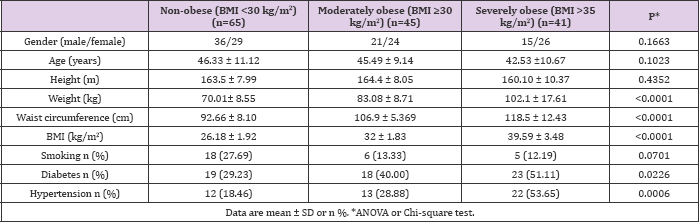
The UCP2 45-bp insertion (I)/deletion (D) genotypes D/D, I/D, and I/I were detected to occur with a frequency of 58.3%, 36.4%, and 5.3% across the total study population, respectively, while the deletion and insertion alleles were detected in 76.4% and 23.6% of study subjects, respectively. The D/D genotype was found to be more highly prevalent in the severely obese group (82.9%) compared to the non-obese (46.2%) and moderately obese (53.3 %) groups. Table 2: Association of the UCP2 54-bp insertion (I)/deletion (D) The conducted logistic regression analysis showed no significant association between the I/X genotypes (where X represents a nonspecified allele) and the development of moderate obesity (OR = 0.75, 95% CI: 0.35-1.59, P = 0.585); however, a strong inverse association was found between these genotypes and severe obesity (OR= 0.18, 95% CI: 0.07-0.44, P = 0.0004) (Table 2).
Table 2: Association of the UCP2 54-bp insertion (I)/deletion (D) polymorphisms with obesity.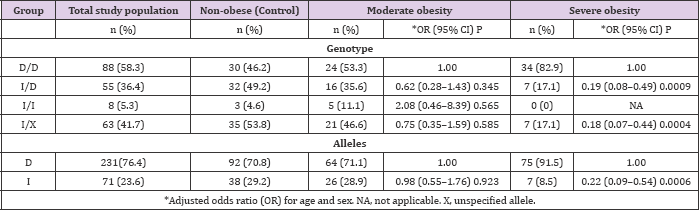
The role of the 45-bp insertion/deletion polymorphism in exon 8 UCP2 gene in the development of obesity is to date, unclear. Thus, the current study investigated the frequency of this polymorphism in groups of non-obese, moderately obese, and severely obese subjects originating from the eastern province of Saudi Arabia. While no significant association was found between either the insertion or deletion alleles and moderate obesity (Table 2), nor either an 'overweight' BMI (25-30 kg/m2) or type II diabetes mellitus (data not shown), a strong association was identified between the D/D genotype and severe obesity in this population. In fact, subjects with this genotype were found to experience a fivetime greater risk for developing severe obesity that than those who harbored at least one insertion allele. The majority of previously conducted association studies have reportedly found no direct link between the UCP2 45-bp insertion/deletion polymorphism and either obesity or various related metabolic disorders [13-18]. Notably, some previous studies showed the insertion allele to be positively associated with obesity, contradictory to the findings of the present study [19-22]. Nevertheless, various studies have produced results consistent with those presented here, suggesting that the D/D genotype may contribute to the development of obesity and its related metabolic disorders.
For instance, the deletion allele has been previously associated with lower resting metabolic and energy expenditure rates in young adults, and thus postulated to contribute to the development of obesity at a later age [8,23]. Similarly, subjects carrying the insertion allele in a (metabolically healthy) Greek population were previously shown to exhibit improved weight loss profiles compared to those harboring the D/D genotype [18]. Previous screening of a Tongan population with a known increasing prevalence of obesity revealed the D/D genotype to be highly prevalent (97%) [24]. Finally, a recent study in the region adjacent to the population analyzed by the present study reported a significant association between the D/D deletion and metabolic syndrome, for which obesity is a well- known risk factor [12]. In Saudi Arabia, obesity is a common health concern caused by excessive caloric intake and/or low physical activity levels. The reduced metabolic and energy expenditure rates incurred by the D/D genotype likely aggravate the tendency to develop severe obesity in this population [25].
Notably, previous analysis of a western Saudi Arabian population showed the insertion/deletion polymorphism to be neither associated with obesity nor type II diabetes mellitus [26]. This discrepancy may reflect the fact that this previous study did not consider moderately and severely obese subjects separately, or alternatively, may reflect genetic differences incurred by geographical separation of the two populations. In conclusion, the present study showed the distribution of the UCP2 D/D, D/I, and I/I genotypes in the analyzed eastern Saudi Arabian population to be 58.3%, 36.4%, and 5.3%, respectively. Moreover, the D/D genotype was shown to be strongly associated with severe, but not moderate obesity. These finding provide valuable insights into the genetic contribution of UCP2 to obesity and related disorders in Saudi Arabia, and should be confirmed via additional research with larger and more geographically diverse cohorts.
I take this opportunity to thank Mrs Amani Al Omairy for her great support to conduct this research.


