Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Sunil Pandey, Laina Sallam and Nivedita Pareek
Received: October 06, 2017; Published: October 17, 2017
Corresponding author: Sunil Pandey, Department of Biological sciences, Eastern Illinois University, Charleston, USA
DOI: 10.26717/BJSTR.2017.01.000446
Staphylococcus aureus is a potentially pathogenic bacterium that causes a wide range of diseases. These are causing different infections and resistance mechanism due to of its wide virulence factors. The increase in the resistance of this virulent pathogen to antibiotic, day by day increases as a nosocomial infection is a major health concern. The main resistance phenotype associated with the S. aureus in hospital is methicillin resistance followed by vancomycin resistance. Resistance to methicillin and other b-lactam antibiotics is produced by the mecA gene, which is located on a portable genetic element, the Staphylococcal Cassette Chromosome mec (SCCmec). Center for disease control and prevention (CDC) approximates 80,461 hostile Methicillin resistance staphylococcal (MRSA) infections and 11,285 associated deaths occurred in 2011.
Methicillin resistance in hospital acquired infections in S. aureus isolates has been increasing intensely in United States and occurring resistance to other antibacterial compounds. However, the role of evolutionary change in the pathogen throughout the development from bearing to disease is not completely understood. So, for this high throughout genome sequencing project need to be done to identify the genotypic character. To reduce these resistances more effective infection control, developing the new and improved antibiotic, developing vaccines, monitoring the trends in resistance, promoting interventions, conducting research are crucial. This review labels the latest molecular evolution of MRSA, different methods used to investigate the epidemiology, different risk factor associated with it and the structure of gene.
Keywords: MRSA; Evolution; Review; Staphylococcus aureus
Abbreviations: SCCmec: Staphylococcal Cassette Chromosome Mec; CDC: Center for Disease Control; MRSA: Methicillin resistance staphylococcal; VRSA: Vancomycin Resistant Staphylococcus aureus; PBP: Penicillin-Binding Protein; UPGMA: Unweighted Pair-Group Matching Analysis; MLST : Multilocus Sequence Typing; SCVs: Small Colony Variants
Nowadays, one can classify infections based on settings in which people acquire them. Precisely, the hospital corresponds to pathogens that possess multidrug-resistant genes whereas community-acquired strains contain virulent genes. However, treating infections with antibiotics can change the entire status. For instance, in the forties of the past century, treatment tuberculosis with streptomycin was enough to refer the disease to the curable ones. Nevertheless, humankind did not exert adequate effort to eradicate the infection completely, and its long-lasting treatment with antibiotics resulted in developing the resistance on the part of the bacteria. Unfortunately, currently, tuberculosis is an incurable disease. Experts expect that further heavy reliance on antibiotics can lead to converging with the virulent gene on the part of the multidrug-resistant one. Such the implies the appearance of pathogens that will require devising new approaches to coping with the several outbreaks. Given this threat, humankind will have to decline the use of antibiotics at some point. Therefore, one can expect that the research in the field has a short time horizon. Therefore, funding, it is limited. For example, in six chosen statistical studies of different aspects of treating infections with antibiotics the used samples are not large enough to guarantee the acceptable level of uncertainty. This report presents the reviews of the many articles that are mainly studied Staphylococcus aureus. S. aureus is a shared type of bacteria that is found on the skin.
Also, found on the catheters, ventilators or patients who Int for surgical procedures, can pass in the body and cause infections. MRSA is related with enlarged mortality and lengthy hospital stays producing important economic expenses. Antimicrobial resistance is worldwide problem. In humans and animal’s pathogens is growing day by day and creating a problem and have incomplete the real lifespan of newly developed antimicrobial up to only 10- 20 years. World health organization have stated antibiotic resistant microorganisms as “nightmare bacteria” that “pose a catastrophic threat” to the world population [1]. CDC estimates 80,461 invasive Methicillin resistant Staphylococcus aureus (MRSA) infections and 11,285 related deaths occurred in 2011. Since 2002, 17 cases of vancomycin resistant Staphylococcus aureus (VRSA) have been identified [1].
S. aureus strains have established resistance to methicillin and vancomycin through the obtaining of the mecA and vanA genes [2]. Penicillin and its different products, including methicillin, vancomycin have been used for the actions of infections caused by S. aureus [3]. However, certain strains of S. aureus developed resistance known as methicillin resistant Staphylococcus aureus (MRSA)As rapidly new antibiotics are introduced in market S. aureus are showing mechanisms to neutralize them [2,3] and this the diversity of the S. aureus is largely determined by the incidence of mobile genetic elements, which consist of prophages or phagerelated genomic islands. Different strain evolution and horizontal gene transfer are closely linked to phages [3]. The purpose of this review is to study evolution of antimicrobial resistance in Staphylococcus aureus.
Different treatment methods and protocol and mechanism of resistance, Multiple mutations lead to the evolution of antimicrobial resistance not just only with single base pair mutations and evolution is also affected by chance not only with the selection [3-5]. By many original and another standard database and were analyzed focusing the MRSA and VRSA. As we have seen that many people confronted with the maximum serious problems in relation to appearance of antimicrobial resistance in S. aureus. We also believe this short review will help to investigate further laboratory experiments and based on the reference available and by combining new data that will be used for further research and review.
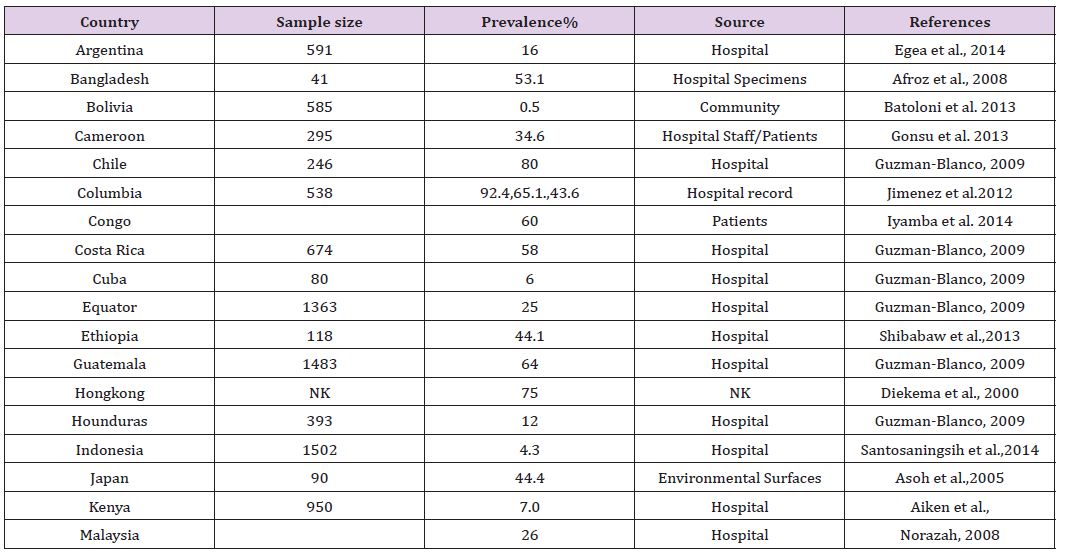
MRSA is caused by the presence of mecA gene. The geen encodes for 78-kDa penicillin-binding protein (PBP) 2a or (PBP2’) [6]. Afterwards mechanism in methicillin resistance was described in S. aureus by Porrero et al. 2014, García-Álvarez et al. 2011, and Paterson et al. 2012. Harrison et al. 2013 suggested the public health hazard of mecC-positive MRSA isolates as it has been isolated in human case and their livestock. The reported cases of community acquired-MRSA infection in the US were caused by a USA400 strain, MW2 [7,8]. USA400 has been supplanted by USA300, which is certainly the most recurrent cause of community acquired-MRSA infections in the US [9]. The infections in the community and veterinary species are in increasing concern [10]. Proper knowledge on the epidemiology of MRSA will support actual anticipation and control strategies, counting the rational use of antibiotics (Table 1).
Table 1 : Prevalence of MRSA in some countries.
According to CDC report a total of 13 cases of vancomycinresistant Staphylococcus aureus (VRSA) have been identified in the United States since 2002 and these are risen from MRSA. As compare to MRSA, VRSA occurrence is low. In the world, the total number of VRSA appealed to be rarer than other bacterial resistance [11]. All VRSA recognized in the United States seem to have autonomously developed [12] the vanA operon. VRSA arise from highly transmissible MRSA progenitor strains, and a vigorous public health response to all reported VRSA is suggested. VRSA strains gain resistance by conjugal transfer of the vanA operon from an Enterococcus faecalis, raising the threat of a far more effectual means for distribution of the resistance gene between strains of staphylococci [13, 14].
Pseudomonas aeruginosa and Staphylococcus aureus often infect pediatric patients simultaneously. The authors selected 33 isolates of the first bacteria obtained from patients for whom the antibiotic treatment was successful, and 13 isolates of this pathogen obtained from people for whom the outcome of the treatment was negative. The researchers studied the biofilm growth using the microscope and the 3D visualization software. They discovered that Staphylococcus aureus determines the structure of the biofilm Pseudomonas aeruginosa form. Such influence is the mechanism of resisting to antibiotics on the part of Pseudomonas aeruginosa isolates [15]. That same flaw is also exploring epidemic behavior of methilicin-resistant Staphylococcus aureus (MRSA) [16].
The use of pulsed-field gel electrophoresis provided the opportunity to register isolates of MRSA that take place only on rare occasions and have genomic DNA patterns that differ drastically from the ones of main clones of MRSA capable of initiating an epidemic. In the Hallin, et al. [15], the researchers name them as the sporadic isolates [15,16]. Using the sample that contains 36 MRSA isolates, the research team identifies four distinct classes of the sporadic ones: The isolates with evolutionary history drastically different from healthcare associated MRSA clones and some characteristics of virulent MRSA strains; Isolates that have an ancestor susceptible to methicillin from whom epidemic isolates were derived, and an apparent type of staphylococcal cassette chromosome mec (SCCmec); Isolates that have the evolutionary history identical to the one the epidemic strains possess or are their closest descendants; Isolates that illustrate the transition from the ancestor of epidemic strains to them. Measuring growth rates artificially informs that sporadic isolates do not lag behind the epidemic ones.
Hence, determining the type of SCCmec and manipulating with various mobile genetic elements one can modulate the ability of MRSA strains to cause epidemics without exerting influence on the fitness cost. The existence of virulent MRSA has to raise concerns since treating it with antibiotics can result in an appearance of superbug with converged virulent and multidrug-resistant genes. Therefore, I believe applying susceptibility testing to sporadic isolates that contain both types of genes introduce unjustified risk. Marchese et al. 2009 illustrate this claim.
TTo prevent the spread of MRSA and VRSA it is requiring the knowledge of both the laboratory dissemination and epidemiology of MRSA strains. For this purpose, various molecular typing techniques have been developed. Pulse filed gel electrophoresis is still measured to be the reference standard for typing MRSA isolates [17], it is also the demonstrated as one of the best most typing methods for studying nosocomial infections. These are based on the based-on digestion of purified chromosomal DNA with restriction enzyme SmaI, followed by agarose gel electrophoresis [18]. The patterns are analyzed through dice coefficient and unweighted pair-group matching analysis (UPGMA) settings [19]. Multilocus sequence typing (MLST) excellent tool for investigating the clonal evolution [20]
MLST is created on sequence analysis of 0.5-kb fragments from seven S. aureus housekeeping genes, i.e., arcC, aroE, glpF, gmk, pta, tpi and yqiL [20]. Four approaches are presently accessible for the classification of SCCmec. Oliveira and de Lencastre [21] developed a multiplex PCR for SCCmec type’s I–IV, in which mecA and six different loci on SCCmec are detected. [22] Established a singlelocus sequence typing method for S. aureus using the sequences of the polymorphic region X of the S. aureus protein A (spa) gene. The main benefit of spa typing over MLST is its easiness; meanwhile it includes sequencing only a single locus.
MRSA arose inside 2 years of the overview of methicillin in 1959. These strains, which concealed SCCmec type I, were isolated in the UK. During the 1960s, MRSA strains were isolated in other European countries, and then during the 1970s from other parts of the world [21] as a cause of nosocomial infection worldwide. A study of 147 MRSA isolates with geologically different origins showed that MRSA has emerged at least 20 times subsequent attainment of SCCmec, and that the gain of SCCmec by MSSA was four-fold more mutual than the spare of one SCCmec with another [23,24]. Interestingly, SCCmec type IV was found in twice as many MRSA clones as other SCCmec types [21,24,25]. Which suggest most clones arise by most clones arise by acquisition of SCCmec type IV by S. aureus [26,27].
Staphylococcus aureus can form small colony variants (SCVs) which makes treating the infection be cumbersome. The Cao S et al. [28] paper sheds light on the matter. The existence of SCVs makes the treatment of the infection due to Staphylococcus aureus be extremely challenging since they are resistant to antibiotics. However, previous studies claimed that SCVs tend to lose the resistance thanks to the compensatory mutation. The study takes advantage of selection with serial passaging to investigate the process. They use the sample that contains 107 SCVs resistant to kanamycin and discovers four alternatives of progressing on the part of the mutation. For the first time, researchers detect the alternative that does not result in the loss of the resistance.
It corresponds to the situation, in which the bacteria produce the abnormally large amount of adenosine triphosphate due to alternative transcription. This chemical accelerates the growth. It turns out that in this case, the microorganisms remain resistant to amino glycoside antibiotics. As for other detected alternatives, in all of them, the bacteria lose the resistance [29,30]. In another study, Daniel et al. [31] Describes 26 types of such the antibiotics in terms of the sites of the interference, its process, and the resistance mechanism. Although such the review provides the input valuable for devising new antibiotics, exerting the impact on protein synthesis genes can converge. Experts expect that further heavy reliance on antibiotics can lead to converging with the virulent gene on the part of the multidrug-resistant one. Given this threat, humankind will have to decline the use of antibiotics at some point.
In the past, people acquired MRSA only in a hospital setting. However, nowadays this bacterium is virulent. The research in hand aims at determining the properties of MRSA strains, genetic background, and resistance to antibiotics. The occurrence of MRSA segregation from hospitals, community has amplified in so many topographical places as compared with VRSA. The continuous care of MRSA and VRSA through monitoring will help in effective control. Understanding of the forces that direct the evolution of virulent and drug-resistant organisms also helps to identify the problems associated with it to fight on timely manner, overuse and misuse of antibiotics is clearly a contributing factor for resistance. Different New technologies principal to enhance and more rapid diagnostics, an improved understanding of pathogenesis of staphylococcal disease, and non-antimicrobial approaches to prevention and treatment of infection will also be needed to anticipate the coming of the post-antibiotic age.


