Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Giuseppe Manco*
Received: September 29, 2017; Published: October 10, 2017
Corresponding author: Giuseppe Manco, Istituto di Biochimica delle Proteine, Consiglio Nazionale delle Ricerche, Via P. Castellino 111, 80131, Napoli, Italia
DOI: 10.26717/BJSTR.2017.01.000424
It is well established that oxidative stress from mitochondria plays an important role in apoptosis and also leads to premature aging and cancer [1]. There is growing scientific consensus that proteins with antioxidative functions, such as paraoxonases, can lower the incidence of these diseases. The paraoxonase (PON) gene family consists of three members: PON1, PON2, and PON3. PON genes are located in a gene cluster on chromosome 7q21.3-22.1 in human [2] and show about 70% sequence identity among them [3]. PON2 is a calcium-dependent glycoprotein of about 44 kDa, expressed ubiquitously, and associated with plasma membrane fractions. Recently, it has been demonstrated that PON2 is a type II Tran membrane protein, with its N-terminal region identified as a single Transmembrane domain, whereas the catalytic domain, corresponds to the C-terminus, located extracellularly to counteract lipid per oxidation [4].
PON2 has two main activities: a calcium-dependent hydrolytic activity, involved mainly in the hydrolysis of lactones, esters and aryl esters [2] and a redox function, which reduces the levels of ROS (reactive oxygen species) thus curbing cell oxidative stress and therefore displaying an anti-apoptotic effect. No clues are available about the molecular basis of this anti-ROS activity except the demonstration that PON2 can bind ubiquinone (coenzyme Q10) with high affinity only in the presence of Ca2+ [5]. In doing so coenzyme Q10 is substracted from interaction with O2 and production of ROS.
Unlike PON1 and PON3, PON2 is an intracellular protein, expressed in a wide range of cell types, including pancreatic beta cells. Importantly, polymorphisms in the PON2 protein (S311C and A148G) have been associated by several studies with diabetes and its complications. Although a genetic association between PON2 and these conditions has been shown, the exact function of PON2 in humans is not known. It has been shown that all of the PONs can inactivate the lactone 3-Oxo-C12-HSL [2,6-8]. Among PONs, PON2 has the greatest lactonase activity against the bacterial quorumsensing (QS) molecules [2,9]. QS Is an interbacterial mode of communication accomplished through the coordinated production, secretion, and detection of chemical signals (QS signals) that trigger the expression of specific bacterial genes. The QS signals self-produced by P. aeruginosa are in the form of small molecules, termed acyl- homoserine lactones (acyl-HSLs) [10] among which 3-Oxo-C12-HSL is the master regulator [11]. It has been demonstrated that PON2 responds to the bacterial quorormone 3-Oxo-C12-acyl homoserine lactone by a rapid decrease of the lactonase activity via a putative post-translational modification (PTM) [12]. Therefore, Pseudomonas produced 3-Oxo-C12-HSL, organizes from one side the coordinated attack of bacterial cells against human tissues and on the other curbs the human defences by blocking PON2 activity, which is thought to represent the first line of defense against infections [9].
PON2 being located on cell surface [4] can rapidly clear out the 3-Oxo-C12-HSL accumulating outside the cell and passing through the membrane. The increase in the concentration of the quorormone activates, at a certain threshold, the production of virulence factors and the formation of a biofilm [11]. It also diffuses inside the cells or binds specific receptors thereby activating processes that are different in different cell types [13]. Relevant is the effect on NFkB signaling through which 3-Oxo-C12-HSL attenuates the innate immune system to establish and maintain local persistent infection in humans, for example, in cystic fibrosis patients [14].
Recently we discovered a new PTM of PON2 induced by 3-Oxo-C12-HSL [15] namely the ubiquitination of Lys 144 (168 in the paper) that may be of physiological importance because it decreases the PON2 activity. Moreover we have shown that PON2 is able to interfere efficiently with the formation of the biofilm in vitro [15]. These experiments were conducted by expressing an engineered version of PON2 in E. coli and renaturing it from inclusion bodies. We also suggested in this way that glycosylation of PON2 is not important for activity but likely for stability. Teiber [16] reported that phosphorylation of Ser36 is necessary for activity. This is in contrast with our data: our rPON2 made in E. coli is active and devoid of phosphorylation also because the deletion at the N-terminus includes Ser36. In any case 3-Oxo-C12-HSL does not seem to act via this phosphorylation site [16].
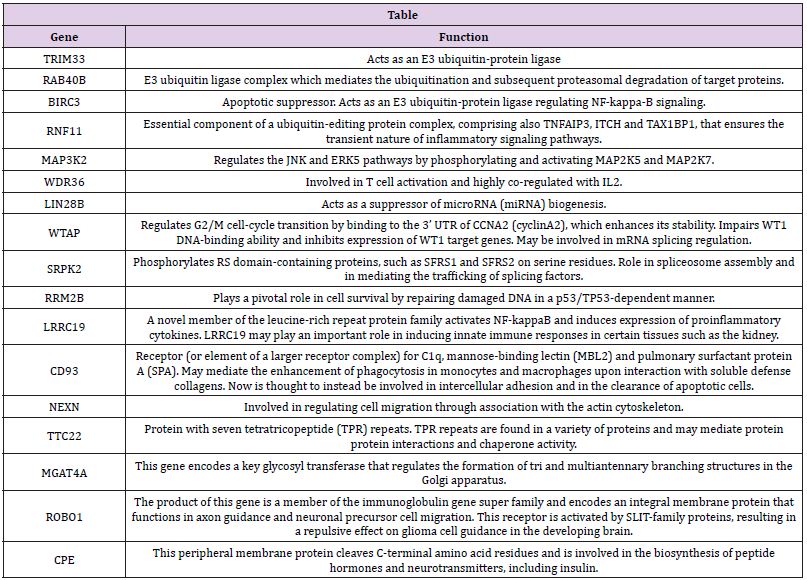
In previously published large scale mass experiments Tyr346 has been found phosphorylated [17]. Mutants at that position maintain activity and are still inactivated by 3-Oxo-C12-HSL [16]. In our effort to clarify the role of PON2 we are focusing also on expression regulation. In a follows up of our previous work [18] we identified in the 3’UTRs of some MHCII genes a quite well conserved sequence, already suggested in a recent paper as a putative regulative signal [19]. The sequence was shared by a cluster of a limited group of genes. In the same database we looked for the presence of signals downstream of the PON2 gene. There was just one occurrence for PON2. The identified sequence was a 12mer: TTTCCTTAAAAT identifying a cluster of 21 genes. This sequence does not overlap with any MREs. The interesting aspect of this finding is that most of these genes are functionally related (based on information at www.genecards.org) and involved in apoptosis, proteosomal degradation (ubiquitinylation) and DNA repair (Table 1). We are currently analysing the involvement of these genes in regulation and physiology of PON2.
Table 1:
This work has been supported by CNR project “Interomics” to G.M.


