Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Sajjad Ullah1,2*, Fayyaz Ahmad2, Ahmad Bilal Waqar2, Arshad Jamal2 and Muhammad Idrees1,3
Received: August 23, 2017; Published: September 08, 2017
Corresponding author: Sajjad Ullah, Centre for Applied Molecular Biology, 87-West Canal Bank Road Thokar Niaz Baig, University of the Punjab, Lahore, Pakistan
DOI: 10.26717/BJSTR.2017.01.000340
Background: Hepatitis C is rapidly emerging as a major health problem in developing countries including Pakistan that leads to death and morbidities. HCV has a high genetic variation and is classified into six major genotypes and 67 subtypes. (Direct-Acting-Antiviral) anti-HCV drugs therapy response, resistance and recovery rates depend on HCV genotype.
Methods: To study the epidemiological study of HCV genotypes in 2015-16 and their main routs of infection in the Punjab Pakistan. Observational study of the patients from 27 centers, 4823 samples was collected from HCV positive patients. History was noted on pre-design tested questioner which was approved by institutional Ethical committee. RNA was extracted using Favorgen Viral Nucleic Acid Extraction Kit. Amplification was performed using Amplisense Genotyping kit. We studied source of infection and therapeutic responses of varies genotypes on the basis of geographical distribution; we found a new incitement and provoke.
Results: A total of 4823 serum samples were tested by type-specific genotyping assay. RNA was extracted using FoverGen Mini Kit. For HCV genotyping AmpliSens® HCV-genotype-FRT PCR kit variant FRT-g1-6. Detail history of each patient was taken on a predesigned questioner which was approved by Institutional Ethical Committee. Total 7800 individuals were analyzed by anti HCV ELISA out of which 5451 patient were found reactive. The positive samples were further conformed by PCR for HCV and their genotypes, out of which 4823 (88.47%) were found detected for HCV RNA. In the division of genotypes in Punjab varies from a maximum of 57.6% the genotype 3a, followed by 3b 14.76% on the other hand least common genotype was type-5 (0.14%). The major route of infection was surgery/dental procedures (52.02%), use of unsafe syringes (18.45%), blood transfusion (16.26%), razors or circumcision (5.90%), less than 3% due to needle stick, while 6.35% was unclear.
Conclusion: HCV The most spread genotype in Pakistan was 3a with rate of 58% followed by genotype 3b and 1a, respectively. Dental surgery was the main source of infection.
Keywords: Hepatitis C Virus; Genotypes; Molecular Epidemiology; Risk Factor
Abbreviations: EDTA: Ethylene Diamine Tetra Acetate; PCR: Polymerase Chain Reaction; RNA: Ribonucleic Acid
Hepatitis C is rapidly emerging as a major health problem in developing countries including Pakistan [1,2]. Prevalence of HCV may be different in different regions and various groups of the same community [3]. Hospital-based studies on HCV revealed prevalence rates of 5.31% (Islamabad), 2.45% (Rawalpindi), 4.06% (Multan), 20.89% (Faisalabad), 4 - 6% (Karachi), 9% (Mardan), 5% (Buner, NWFP) and 25.7% (Northern Areas) [4]. Slightly higher prevalence of HCV was recorded in the earth quake effect areas of Pakistan in 2005 [2]. A recent study showed that in Pakistan more than 90% of HCV positive subjects were unaware of HCV infection in general [3]. Along with host factors such as age older than 40 years, female sex, and low baseline viral load, the less number of mutations in the HCV-3a is associated with a positive outcome of treatment response in HCV patients receiving interferon plus ribavirin therapy [5].
It has been observed that the prevalence of HCV increases with age which may be due to increasing exposure to risk factors and, the most prevalent genotype in Pakistan was 3a with rate of 50% followed by genotype 3b and 1a, respectively [6]. In this study we aimed to determined extensive information on genotypes and their sub types distribution based on available patients from different geographical regions of routinely diagnosing and treating HCV-infected patients also linked the HCV distributions through different parameters. Additionally, we have find the source and rout of hepatitis C infection.
Samples were collected from 27 different centers of the observed patients who had serum HCV RNA levels of 10,000 IU per milliliter or higher from patients who had never received any type of treatment for HCV infection. A detailed history was taken on the pre-design tested questioner which was approved by institutional Ethical committee. Viral RNA isolation was carried out using Favorgen Viral Nucleic Acid Extraction Kit (Favorgen® Biotech Corp) according to the manufacturer’s instructions. Reverse Transcription PCR (RT-PCR) was used for the synthesis of cDNA using Amplisense Genotyping kit. The amplification and genotyping were performed by using the manufacturer instruction. We studied source of infection and therapeutic responses of varies genotypes on the basis of geographical distribution; we found a new incitement and provoke.
The study was conducted to explain the distribution of HCV genotypes and their main causes of spreading in the population of Punjab, Pakistan. Seven Thousand and eight hundred patient were checked by anti HCV ELISA out of which five thousand four hundred and fifty one patient were found reactive which was further proceeded by HCV PCR, simultaneously serum samples, in 4823 (88.47%) serum samples the HCV RNA were detected. Three ml blood sample was collected from 5451 patients (HCV reactive by ELISA); in ethylene diamine tetra-acetate (EDTA) tube. Plasma was obtained by centrifugation of blood at 600 rpm for 15 minutes. RNA was extracted using Favorgen RNA extraction kit. HCV genotyping by using For HCV genotyping AmpliSens® HCV-genotype-FRT PCR kit variant FRT-g1-6 by real time PCR the kit was planned to detect HCV major 6 genotypes with their sub types. Detection of HCV genotypes 1a, 1b, 2, 3a, 4, 5a and 6 by polymerase chain reaction (PCR) is based on the amplification of a pathogen genome specific region using specific primers. In real-time PCR, the amplified product is detected by using fluorescent dyes. These dyes are linked to oligonucleotide probes which bind specifically to the amplified product.
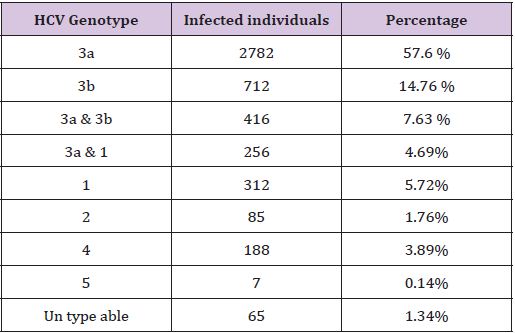
Table 1: The Distribution of HCV Genotypes in the Punjab Pakistan.
The screening of blood for anti-HCV is mandatory seven thousand and eight hundred patient were checked by anti HCV ELISA, thus the patient who were found to be positive for said infections then the serum samples were tested for HCV by ELISA. The ELISA positive samples were further processed for more confirmation on PCR, out of total (Table 1), total 4823 people affected with HCV out of which 67% and 33%, male and female respectively. As regard the genotyped samples, patients were belonging to different areas of the Punjab, and have different geographical variation. Genotype 3a is common in Pakistan. The percentage ratio of genotypes in Punjab varies from a maximum of 57.6% the genotype 3a, to a minimum of 0.14% the genotype 5 (Table 1).
Genotype 3b and 1 were distributed 14.76% and 5.72% respectively. Some of the patients have a mixture of genotypes. The most common combination is 3a and 3b. In some patients the genotype was unable to type. The main route of HCV transmission was major/minor surgery/dental procedures (52.02%). Intravenous drug abuse and syringes (18.45%), blood transfusion and blood products (16.26%), sharing razors or circumcision (5.90%), piercing instruments, less than 3% due to needle stick. For about 6.35% subjects the mode of transmission was unclear.
According to a recent study, in Pakistan the observed genotypic distribution were 3a (49.05%), 3b (17.66%), 1a (8.35%), 2a (7.52%), 1b (3.01%), 4 (1.49%), 3c (0.75%), 2b (0.80%), 1c (0.15%), 6a (0.12%), 2c (0.09%) and mixed infection (4.80%) [6]. The detection of HCV RNA by reverse transcriptase PCR has become essential for the diagnosis of HCV infection and for the selection of patients before therapy. The main advantages of reverse transcriptase PCR include early diagnosis after acute infection and detection of viremia in selected patients (those with indeterminate antibody results and immune suppressed patients). The sensitivity of PCR for HCV RNA detection may vary according to the choice of primers and the handling of pre-extraction samples [7,8].
The study was conducted to explain the distribution of HCV genotypes and their course of distribution in the population of Punjab. Total 4823 people affected with HCV, genotype 3a was most common type in Pakistan. The percentage ratio of genotypes in Punjab varies from a maximum of 57.6% the genotype 3a, to a minimum of 0.14% the genotype 5 (Table 1), 3b and type 1 were distributed 14.76% and 5.72% respectively. Some of the patients have a mix type which was almost of type 3a and 3b. In some patients it was difficult to define the genotype because it represents a new and unclassified genotype. The mixed genotypes were found in thalassaemic and dialysis patients. Because of geographic grouping of distinct HCV genotypes, beside the treatment duration, genotyping may be a helpful implement for tracing the cause of an HCV epidemic in a given population.
To fine out the new possible routes for HCV transmission patient’s sample along with detailed history of patient was taken. Patient consent was noted. History included Name, age, gender, address, blood donation or transfusion history, operation history, family history, occupation, dental procedure history, nose and ear piercing history, tattooing history and history of reuse of needles for injection was noted. Patients were asked various questions in order to find out the risk factors involved in transmission of HCV (52.02%). Patients gave a history of operations (major or minor) including procedures after road accidents and patients had dental procedures, 32 (5.73%) had HCV positive family members, (18.45%) had donated or received a blood transfusion.
Among patients who had received blood transfusions children suffering from hemophilia and thalassemia were also included. (5.90%) patients gave a history of visiting barber shops for shaving and less than 3% due to needle stick, from infected mother to baby and sexual transmission. For about 6.35% subjects the mode of transmission was unclear. The history mentioned by these patients led to the conclusion that more than one factor may contribute towards the spread of HCV infection. But almost all patients gave a history of dental procedure single or along with other factors. Use of non sterile equipments in dental procedures is playing a vital role in transmission of HCV infection. We diagnose for the very first time 9 different HCV genotypes from Punjab Pakistan. The most common genotype in Punjab was type 3a. The major route of transmission of HCV was surgery and dental procedures.
This research was supported by the Higher Education Commission of Pakistan.
This study was partially supported by Higher Education Commission of Pakistan.


