Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Alain L. Fymat*
Received: May 22, 2017 Published: June 05, 2017
Corresponding author: Alain L. Fymat, International Institute of Medicine and Science, California, USA
DOI: 10.26717/BJSTR.2017.01.000117
According to the World Health Organization (WHO)’s “First Global Report on Antibiotic Resistance”, and the U.S. Centers for Disease Control & Prevention (CDC&P), the spread of “superbugs” - bacteria that have changed in ways that render antibiotics ineffective against them - is a serious and growing threat around the world. Once common treatments for everyday intestinal and urinary tract infections, pneumonia, infections in newborn, and diseases like gonorrhea are no longer working in people. Thus, in 2013, 2 million people in the U.S. were infected with antibiotic-resistant bacteria, and 23,000 of them die each year as a result.
Superbugs are bacteria-resistant to one or more antibiotics, and they make it difficult to treat or cure infections that once were easily treated. The antibiotic has lost its ability to control or kill bacterial growth. The bacteria can even grow in a sea of antibiotics because the antibiotic does not touch them. The bacteria have acquired the ability to destroy the antibiotic in order to protect themselves. They have developed a gene for resistance to, say, penicillin, and that gene protects them. A genetic mutation might enable bacteria to produce enzymes that inactivate antibiotics. Or, a mutation might eliminate the target that the antibiotic is supposed to attack. Bacteria may have developed resistance to five or six antibiotics so, in treatment, we do not know which one to choose or whether it will be effective. The bacteria have accumulated resistance by developing new genes. In other words, genetics is working against us!
Misuse of antibiotics to treat viruses rather than bacteria for which they are intended only contributes to antibiotic resistance. The same holds true for other uses whereby 80% of the antibiotics produced are fed to animals: beef cattle, chickens, hogs,... to help them grow “better” and put on more weight. The animals excrete the antibiotics in a largely unbroken form, which enter the environment (ground, water) and retain their ability to affect bacteria and promote antibiotic resistance. There is now a paucity of new antibiotics to take care of these superbugs. So we remain at the mercy of bacteria!
Additionally, there is a growing scientific awareness of the impact of antibiotics on the human and animal microbiome - our internal environment of commensal, symbiotic, and pathogenic microorganisms that completely shares our body space, are essential to our gut efficiency, and are at the centre of our immune system. The impact of antibiotics on this internal environment is devastating and links are rapidly being made between our overuse of antibiotics and the epidemic of chronic diseases such as cancer and diabetes. 23,000 deaths a year from superbugs is just a small and obvious part of the story. Much more insidious and pervasive is the wider story of chronic disease that massive overuse of antibiotics is contributing too.
In this paper, to better understand the important phenomenon of antibiotic resistance, I shall begin by a review of antibiotics: their discovery, the history of their development, their modes of action, their clinical activities, and the factors determining response to therapy with antibiotics. I shall then investigate the mechanisms of antibiotic resistance, review some major epidemics due to antibiotic resistance and infectious diseases and antibiotic resistance. Antibiotics do not work all the time, their inappropriate use, nosocomal infections, and possible reservoirs of antibiotic resistant animal organisms causing human diseases will then lead us to an update on the present situation, highlighting the nanopore revolution in genomic sequencing of drug-resistant bacteria, precious nanometals, and look forward to the future of antibiotics.
Abbreviations: AHA/JCAH: American Hospital Association/Joint Commission on Accreditation of Hospitals; ARI: Antibiotic Resistance Island; CDC&P: (U.S.) Centers for Disease Control & Prevention; CRE: Carbapenem-Resistant Enterobacteriaceae (a superbug); ESBL: Extended- Spectrum Beta-Lactamase; FDA: (U.S.) Food & Drug Administration; MDR: Multiple Drug Resistance; MIC: Minimum Inhibitory Concentration; MRSA: Methicillin- Resistant Staphylococcus aureus (MRSA); MSSA: Methicillin-Sensitive Staphylococcus aureus; ORSA: Oxacillin-Resistant Staphylococcus aureus; NAS: (U.S.) National Academy of Sciences; NRC: (U.S.) National Research Council; VRE: Vancomycin-Resistant Enterococcus; T2DM: Type 2 Diabetes Mellitus; WHO: World Health Organization
In 1941, Albert Alexander, a policeman in Oxford, England, was the first patient to ever be treated with an antibiotic. While tending to his garden, he had been scratched by a rose - a scratch that quickly became infected by bacteria, probably Staphylococcus aureus with an admixture of various Streptococci, and turned septic. The sepsis spread, caused the patient’s head to swell with abscesses that necessitated the removal of one eye; in short, he was on the verge of dying. He was treated at the Radcliffe Infirmary by Drs. Howard Florey and Ernst Chain who were brewing up extracts of a mold called Penicillium chrysogenum and had synthesized a very small amount of penicillin. While having been discovered in 1938 by Alexander Fleming, that drug had never actually been used to treat a human. It was effective in improving the policeman’s condition. Unfortunately, not enough penicillin had been synthesized and when the treatment stopped, the sepsis roared back and the patient eventually died. Whereas that story did not end well, millions of other people have since lived because of it, and global health had been transformed.
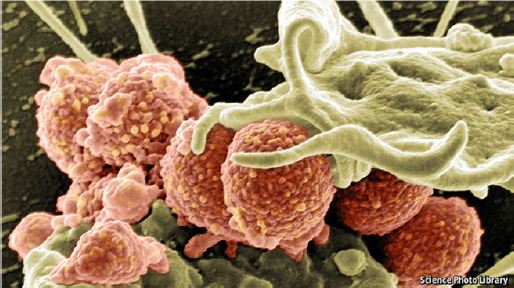
Figure 1: MRSA Being Attacked by a White Blood Cell.
Table 1: The Top Drug-Resistant Threats.
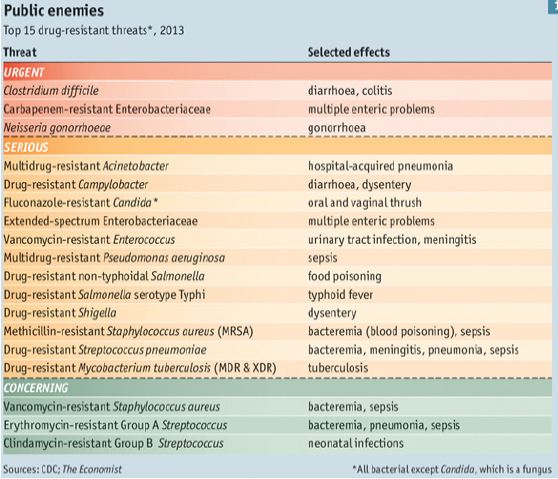
Penicillin is now available in copious amounts, as are other bacteria-killing antibiotics. A study by the CDC&P found that the number of cases of sepsis rose from 621,000 to 1,141,000 between 2000 and 2008, with deaths rising from 154,000 to 207,000. One reason for that is the emergence of methicillin resistant Staphylococcus aureus (MRSA), a variety that cannot be killed with methicillin, one of penicillin’s most effective descendants. Figure 1 shows MRSA being attacked by a white blood cell. This could just be a taste of things to come. Three years ago, the CDC&P produced a list of 18 antibiotic-resistant microbes that threaten the health of Americans (Table 1) in three categories (urgent, serious, and concerning). Five of them (including MRSA) cause sepsis.
Unfortunately, antibiotics have also been used in situations when not really needed and commercially for animal husbandry. That massive use of antibiotics around the world has imposed such large selection pressure on bacteria that resistance has become a serious global problem. Further, resistance has developed to several drugs, including those used for the treatment of tuberculosis, malaria, agricultural pests, which (might) render such treatments ineffective. According to the CDC&P, antibiotic resistance is one of the top five health threats facing Americans. More than 2 million people in the U.S. get infections every year that are resistant to antibiotics, and around 23,000 people die from these infections. Contributing to that problem is the unrestricted use of antibiotics in animal rearing; drugs are used to fatten up livestock and prevent illness, and their routine application has contributed to the rise of so-called “superbugs” (Figure 2) that are resistant to the drugs designed to kill them.
Figure 2: Synthesis of Macrolide Erythromycin-type Antibiotics.
In this paper, I shall begin by a survey of the natural microorganisms that inhabit us and briefly review the history of the development of antibiotic drugs to this date. I will discuss the several modes of action of antibiotics, their clinical activity, and the factors that determine the response to therapy. I will then review the several mechanisms (evolution, spreading) by which microorganisms become resistant to one or several drugs (the so-called “super-bugs”). I will subsequently review the major epidemics of human diseases that were enhanced through the proliferation of antibiotic-resistant bacteria. It is important to note here that antibiotics do not work all the time. I will also discuss the misuses of antibiotics and the global threat they pose to us all. I will briefly address infections caused in hospital settings and the reservoirs of antibiotic-resistant animal organisms that can cause human diseases. Lastly, I will provide an update on the present situation and outline the various treatment failures due to antibiotic resistance. I will conclude on the positive note afforded by the newer nanotechnologies, particularly the so-called “nanopore revolution” in the genomic sequencing of drug-resistant bacteria that could potentially enable bacterial identification, diagnosis of infectious diseases, and detection of drug-resistance at the point of clinical need.
Antibiotics are natural or synthetic chemicals able to affect the survival of microorganisms through inhibiting their growth or killing them. Many molds and mold-like bacteria have rather remarkable bacterial-inhibiting abilities. The modern history of antibiotics devolved as follows:
In recorded history and probably before: Fermenting yeasts and fungi that comprise the sediment in all grain-synthesized alcohol products were the source of many medicinal effects.
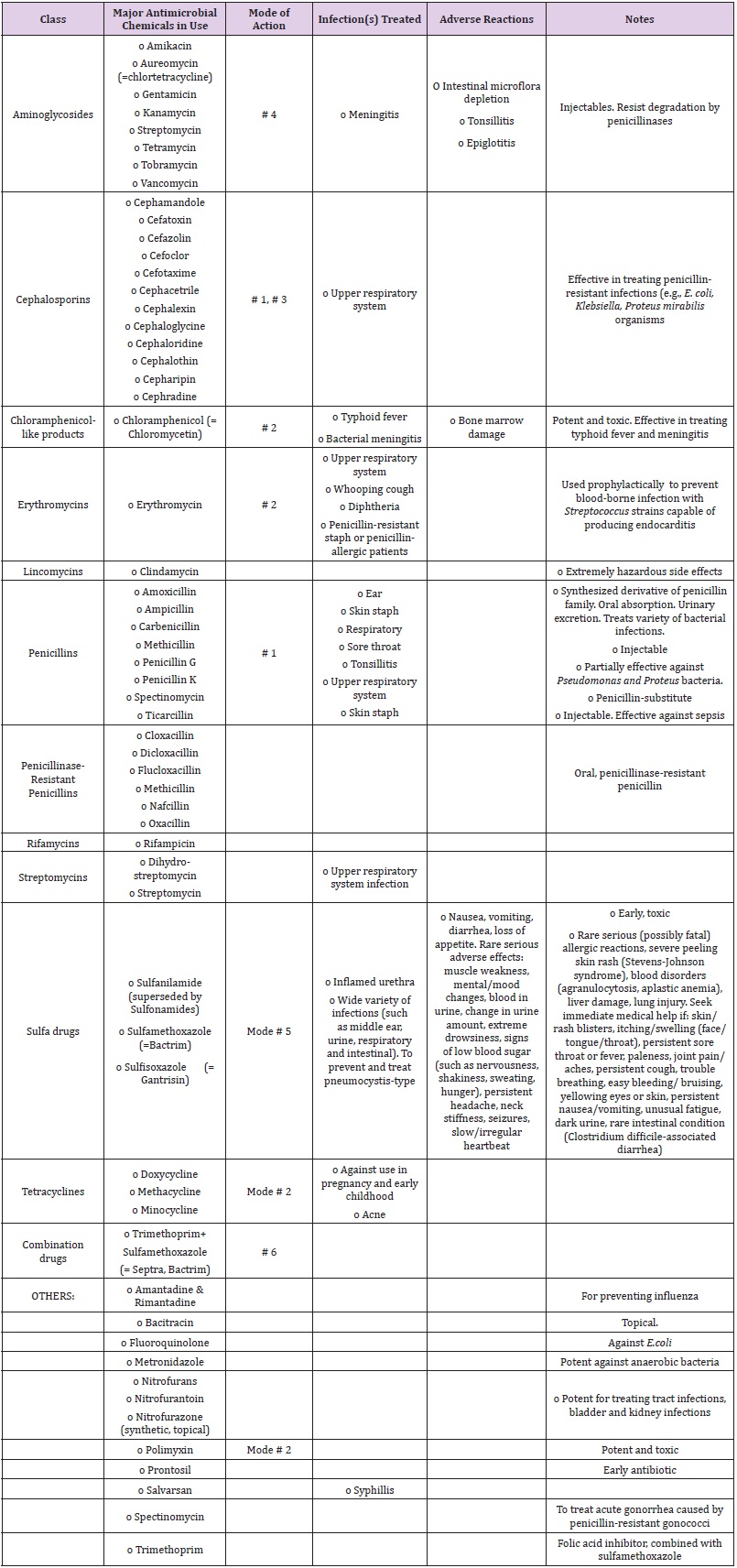
Table 2: Major Antibiotics Currently in Use.
Source: Modified and adapted from D. Perlman “Antibiotics: Old and New”, Wisconsin Pharmacy Extension Bulletin, 18: 1-4, October 1975; and Marc Lappe “Germs That Won’t Die”, Anchor Press, 246 pp, 1982 .
Notes: (Mode # 1) Interfere with cell walls synthesis; (Mode # 2) Inhibit protein synthesis; (Mode # 3) Interfere with protein or membrane synthesis; (Mode # 4) Interfere with the genetic synthesis or operation (DNA or RNA); (Mode # 5) Interfere with the metabolic reactions; (Mode #6) Combination drugs.
Table 2 summarizes the major antibiotics currently in use together with their mode of action, the conditions treated, and their drawbacks and adverse effects. It is worth noting that by 1974, 3,000 different antibiotics and at least 30,000 derivatives have been generated. Creation of new antibiotics continues to this date albeit at a slower pace while only some forty or so are in general clinical practice at any one time (10-15 in Czechoslovakia and China, keeping potent antibiotics like tobramycin “in reserve” for emergencies). It may be noted that over the last 30 years, no major new types of antibiotics have been developed.
The above history continues today as a unilateral quest for chemical rather than homeostatic or immunological solutions advocated earlier by Louis Pasteur and Elie Metchnikoff. These researchers proffered that “natural” antimicrobial chemicals could be discovered in the habitat of microbes themselves, thus relying on the host’s own powers of resistance and immune system. In 2014, a completely new antibiotic named Teixobactin is the latest medicine discovery against multi-resistant organisms. It can kill serious infections in mice without encountering any detectable resistance, offering a potential new way to get ahead of dangerous superbugs such as MRSA/ORSA (methycyllin-resistant Staphylococcus aureus, also called oxacillin-resistant Staphylococcus aureus). MRSA/ORSA is a bacterium responsible for several difficult-to-treat infections in humans. MRSA is any strain of Staphylococcus aureus that, through the process of natural selection, has developed resistance to betalactam antibodies, which include the penicillins and penicillinaseresistant penicillins (methicillin, dicloxacillin, nafcillin, oxacillin, etc.) and the cephalosporins (Table 3). Strains unable to resist these antibiotics are classified as methicillin-sensitive Staphylococcus aureus, or MSSA. The evolution of such resistance does not cause the organism to be more intrinsically virulent than strains of Staphylococcus aureus that have no antibiotic resistance, but resistance does make MRSA infection more difficult to treat with standard types of antibiotics, and thus more dangerous. MRSA is especially troublesome in hospitals, nursing homes and prisons, where patients with open wounds, invasive devices, and weakened immune systems are at greater risk of infection than the general public.
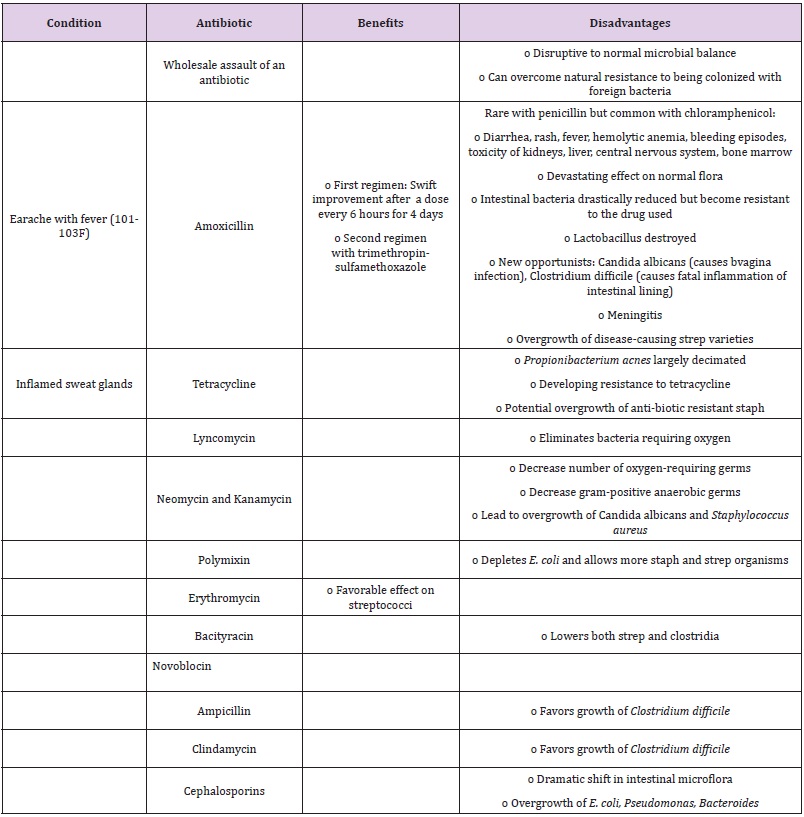
Table 3: Beneficial and Detrimental Effects of Certain Antibiotics.
The teixobactin substance was produced by soil bacteria of mud. Bonn and Boston researchers are at the origin of this discovery (2014). This toxin acts completely differently from all known antibiotics. It attacks the lipid-protective package that has been protecting stubborn bacteria against antibiotics, which includes anthrax and tuberculosis bacilli as well as other highly dangerous pathogens causing pneumonia and blood poisoning. It has yet to be trialled in humans.
The WHO warned last year that a post-antibiotic era, where even basic healthcare becomes dangerous due to risk of infection during routine operations, could come this century unless something drastic is done. Researchers sought to address the problem by tapping into new potential sources of antibiotics. They developed a way of growing uncultured bacteria in their natural environment using a miniature device called an iChip that can isolate and help grow single cells. They have since collected about 50,000 strains of uncultured bacteria and discovered 25 new antibiotics, of which teixobactin is the latest and most interesting one. However, we will not know whether teixobactin will be effective in humans until this research is taken to clinical trials.
A. Note: A gene that confers resistance to a last resort antibiotic has recently [1-3] been detected in the U.S. The gene (mcr-1) decreases the sensitivity of bacteria to colistin, a last resort antibiotic for treating multi-drug resistant infections. It has been found in Escherichia coli and Enterobacteriaceae. While this gene had been found virtually on every continent, it is only now that it has been detected in the U.S. It has been detected at the Walter Reed Army Institute of Research, which has now begun testing all extended-spectrum beta-lactamase (ESBL) that produce E.coli clinical isolates for resistance to colistin [4]. History has shown that mobile resistance genes can spread quickly around the world, riding on people, animals and food. But the news is not unexpected because mcr-1 is contained in a plasmid. The U.S. Is actually an anomaly in not finding it until now. It was probably around for some time, just not detected. This discovery is quite concerning because we would not know what to use in cases of colistin-resistant infections.
For most of the field’s history, natural products have been the starting point for new antibiotics. Most of them have been made by chemically modifying natural products in a process known as “semisynthesis”. Every existing antibiotic in a class called “macrolides”— including the commonly prescribed drug Azithromycin—has been made by modifying erythromycin, which was first discovered in a soil sample in 1949. But, as will be abundantly discussed, some bacteria are developing resistance to these drugs at a startling rate, and semi-synthesis is limited by the difficulty of modifying such complex molecules.
Now, scientists at Harvard University have devised an approach for synthesizing new macrolide antibiotics from simple chemical building blocks [4]. They have synthesized more than 300 new antibiotic candidates, several of which being effective against some of the most stubbornly drug-resistant bacterial strains. This is the first time there is a relatively easy path to synthesize macrolide erythromycin-type antibiotics from scratch (Figure 3).
Figure 3: One Type of “Super Bug”.
The macrolides have been synthesized from eight simple chemical building blocks to produce a diverse collection of structures that would be practically impossible to make using semisynthetic methods. This approach allows for, theoretically, tens of thousands of compounds or more.
Macrolides consist of a macrolactone ring with one or two sugars. “Decorating” these rings with different chemical groups by combining the building blocks in various combinations, Myers’s team created more than 300 synthetic macrolides, including the U.S. Food and Drug Administration (FDA)-approved drug telithromycin and the candidate solithromycin, which is currently being tested in Phase 3 clinical trials.
Next, the researchers evaluated the newly synthesized compounds with a panel of different bacteria, including two strains of MRSA and vancomycin-resistant enterococcus (VRE) isolated from clinical samples. Most of the compounds were effective against garden-variety pneumonia bacteria, and a few of them were more potent than approved antibiotics against MRSA, VRE, and other highly resistant strains. One of the synthetic compounds, called FSM-100573, was especially effective against gram-negative bacteria, which are traditionally less susceptible to macrolides.
These compounds have yet to be tested pre-clinically. And, as with all antibiotics, bacteria will ultimately develop resistance to them. However, this approach allows scientists to develop an exponentially larger number of new drug candidates. This work represents a paradigm shift in the discovery of novel antibiotics from the macrolide class. Previously, Myers and his colleagues have synthesized novel tetracycline antibiotics, one of which is now a Phase 3 clinical trial candidate.
Antibiotics can be distinguished on the basis of their mode(s) of action that is on their ability to interfere with the metabolic machinery of microbes. The microbe needs this machinery to thrive, hold itself together, or make duplicate versions of it. Antibiotics are supposed to thwart bacterial – not human – cells through one or more of the following modes of action:
This is a common requirement for these organisms to cause infections for without the rigid support of the cell wall, most bacteria simply break open and die, or collapse into, an effective heap. Note, however, that a few varieties of bacteria can actually thrive by constructing outer membranes instead of cell walls, but they rarely can reproduce to create further harm. The key molecular target of these antibiotics is a substance (mucopeptide) that coats the bacterial membrane and gives it its rigidity. Examples include such narrow-spectrum antibiotics as:
Penicillins and cephalosporins can inactivate key bacterial enzymes (peptidases) that synthesize the rigid cellular walls, and this all the more effective the thicker the walls. They are remarkably non-toxic at normally used therapeutic dosages because they do not attack human cell walls or membranes which are made from different materials than bacteria.
These are generally more toxic to the human body, and include the broader-spectrum antibiotics such as:
These include:
These include:
Two (or more) chemicals interfere with the same reaction but at different places. Examples include:
All but the most innocuous localized infections initially pit microorganisms against the host’s natural defenses. Antibiotics usually work by keeping invading microorganisms at bay long enough to allow the host to remove and destroy them.
Antibiotic activities are clinically separated between:
However, in practice, an antibacterial drug can be bacteriocidal at a high dose and bacteriostatic at a lower one. Its effectiveness can be determined through two tests:
There are five factors that are considered in evaluating the response to antibiotic therapy:
Below summarizes the beneficial/detrimental effects of certain antibiotics:
Thus, the pervasive use of antibiotics may cause subtle and often imperceptible changes in the microflora with which we have evolved. This is accompanied with the real possibility of indirect havoc to less physically resilient hosts in one’s closer social environment, including the spread of antibiotics tolerance and resistance.
In 2014, the WHO [6] published its first Global Report on Surveillance titled “Antimicrobial Resistance”. In this report, it focused on antibiotic resistance, that is when bacteria change and antibiotics fail, alerting the world that this phenomenon is no longer a prediction for the future but is happening right now across the world. It subsequently published two Fact Sheets titled “Antibiotic Resistance” and “Antimicrobial Resistance”.
Antibiotics work by inhibiting or killing susceptible microorganisms. Among the billions of germs that make up the population of any given infection, a few somehow withstand a low-level antibiotic assault, survive, and if the level of antibiotic remains low or drops, go on to replicate and form a new “antibioticresistant” infection. In other words, bacteria could exist with the innate information needed to resist an antibiotics’ effect even before antibiotics are used.
Resistance could precede the application of antibiotics! It has indeed been demonstrated that chromosomal genes for resistance exist. Antibiotics do not induce resistance (the so-called “induction hypothesis”), as once thought, but rather their inappropriate or/ and inadequate use lead to an evolutionary mechanism wherein microorganisms develop resistance. The antibiotic serves merely to create the conditions that favor the outgrowth of preexisting antibiotic-resistant organisms, and does not induce others to acquire the resistance state. This evolutionary potential of microorganisms (a genetic mechanism) is a key to eradicating the infection if it is incorporated in the therapeutic strategy employed. Evolutionary mechanisms explain both the origins of antibiotic resistance, and the emergence of antibiotic-resistant bacteria.
Further, bacteria spread their resistance information beyond the initial organisms. Populations in far flung regions of the world, who have never known or been treated with antibiotics, or been in contact with people who had been treated with antibiotics, were found to have antibiotics resistance. This demonstrates that resistance is a natural part of the genetic makeup of microbial communities. Information on resistance can be carried on the bacterium’s own chromosome or, when the chromosome breaks off, on these satellites (known as plasmids) leading to two kinds of antibiotic resistance: (a) chromosomal and (b) R-factor mediated. These two mechanisms may be augmented when DNA sequences jump from one DNA molecule to another (called transposons). Transposable resistance may be the most rapid and dangerous form of dissemination of all, since it permits resistance genes to move readily between chromosomes and R-factors and, hence, into the microbial world at large. Further, R-factors can also duplicate themselves within the bacterium itself, making multiple copies of their own chemical instructions thereby augmenting the hazard of antibiotics use.
Further complications are (a) the link between the genes for resistance and those for making some bacteria more virulent, (b) the close physical association between different resistance genes, and (c) the replication of R-factors independently of the bacterial chromosome while inside the bacteria. In other words, the forces that enhance the spread of resistance to one antibiotic simultaneously incorporate resistance to others.
Bacteria resistant to one or more antibiotics have been termed “superbugs” (Figure 3). The WHO defines and describes a superbug thus:
“Antimicrobial resistance occurs when microorganisms such as bacteria, viruses, fungi and parasites change in ways that render the medications used to cure the infections they cause ineffective. When the microorganisms become resistant to most antimicrobials they are often referred to as “superbugs”. This is a major concern because a resistant infection may kill, can spread to others, and imposes huge costs to individuals and society”.
Superbugs make it difficult to treat or cure infections that once were easily treated. The antibiotics have lost their ability to control or kill bacterial growth. The bacteria can grow even in a sea of antibiotics because the antibiotics do not touch them as they have acquired the ability to destroy the antibiotics in order to protect themselves. The bacteria have developed genes for resistance and these genes protect them. Genetic mutations might enable bacteria to produce enzymes that inactivate antibiotics or eliminate the target that the antibiotics are supposed to attack. Further, bacteria may have developed resistance to five or six antibiotics so it is not known which antibiotic to use for treatment. The bacteria can accumulate resistance by developing new genes.
An example of such deadly superbug outbreaks took place beginning in 2009 and lasting to the present date in several hospitals (in Florida, Chicago, Seattle, Los Angeles, Pittsburgh, Hartford, and elsewhere). The cause was contamination from specialized medical scopes, particularly duodenoscopes due to drug-resistant infections. Duodenoscopes are especially difficult to disinfect because of trapped bacteria from a mechanism at the tip of the duodenum. Other causes stem from raw sewage discharged from hospitals, directed to distant treatment plants and released as clear water into water surfaces (streams, oceans,). Unfortunately this treated water is not tested after it flows out of the treatment plant for the presence of the superbug carbapenem-resistant enterobacteriaceae (CRE). This was reported (Los Angeles Times, 7 March 2016) for Southern California hospitals, but was also the case for other U.S. Hospitals. As it turns out, the raw sewage is a highly conducive environment that harbors CRE and allows it to proliferate and grow. As the sewage mixes, the antibiotics kill off weaker bacteria, leaving the more lethal ones to thrive. The bugs reproduce rapidly, and different species can swap genes, transferring their ability to withstand the drugs.
In the U.S., it is estimated that ~ 8% of the number of people sickened with CRE who have not recently visited a medical facility die of it. Hospitals are not breaking laws by releasing the sewage. While laws regulate the overall number of disease-causing bacteria in the nation’s surface waters, there are no specific regulations concerning bacteria-resistant to antibiotics. A 2010 study estimated that 689,000 to 4 million people are struck by gastrointestinal illnesses caught from Southern California beaches each year, and an additional 693,000 are sickened with respiratory problems.
Plasmids may carry genes that provide resistance to naturally occurring antibiotics in a competitive environmental niche, or the proteins produced may act as toxins under similar circumstances, or else allow the organism to utilize particular organic compounds that would be advantageous when nutrients are scarce.
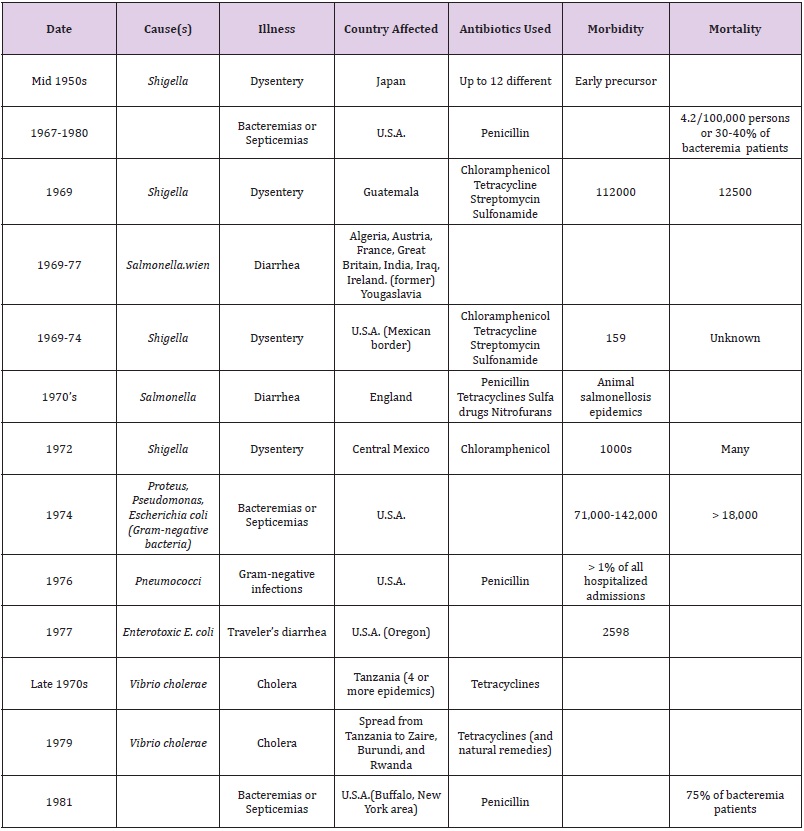
The true extent to which the number and severity of major epidemics of human diseases have been enhanced through the proliferation of antibiotic-resistant bacteria may never be known. Some of these epidemics have been summarized in Table 4.
Table 4:Major Epidemics of Human Diseases Enhanced Through Proliferation of Antibiotic-Resistant Bacteria.
Infections are caused by resistant microorganisms. They often fail to respond to standard treatments resulting in prolonged illness, higher health care expenditures, and a greater risk of death. For serious infections caused by the same common non-resistant bacteria, the death rate for patients can be twice as high when treated in hospitals. On the other hand, consider the case of MRSA, a common source of severe infections in the community and in hospitals. People with MRSA are estimated to be 64% more likely to die than people with a non-resistant form of the infection.
Antimicrobial resistance hampers the control of infectious diseases by reducing the effectiveness of treatment; thus, patients remain infectious for a longer time, increasing the risk of spreading resistant microorganisms to others. When infections become resistant to first-line drugs, more expensive therapies must be used. A longer duration of illness and treatment, often in hospitals, increases health care costs as well as the economic burden on families and societies. And, without effective antimicrobials for prevention and treatment of infections, the success of organ transplantation, cancer chemotherapy, and major surgery would be compromised.
Antibiotics create a kind of closed loop of dependency, first seeming to defeat an infection, while in fact only leaving the host more susceptible to the next bout, thereby creating the need for further antibiotic treatments. These, then, further impair the survival of the inhibiting microbes that would have made the treatment unnecessary in the first place.
In addition, the growth of global trade and travel allows resistant microorganisms to be spread rapidly to distant countries and continents through humans and food. Estimates show that anti-microbial resistance may give rise to losses in Gross Domestic Product of more than 1% and that the indirect costs affecting society may be more than 3 times the direct health care expenditures. It affects developing economies proportionally more than developed ones. But before tackling in detail the problem of antimicrobial resistance, it may be appropriate to first begin with a brief history of the development of antibiotics. Lastly, antibiotics are added to some vaccines to prevent the growth of bacteria during production and storage of the vaccine.
When antibiotics are not used judiciously and with particular attention to the sensibility of the particular organisms being attacked or/and in the presence of foreign bodies or obstructions, they develop new strains that are resistant to particular antibiotics. Likewise, a prior course of antibiotic treatment can alter the patient’s susceptibility to a second infection and end up developing new resistant strains. When antibiotics fail to rescue an infected host, the causes are often multiple, ranging from:
Organisms surviving an initial antibiotic onslaught can pass on their knowledge with dramatic swiftness to another group of organisms, and from there to wider and wider circles of bacterial hosts. Multiple drug resistance (MDR) thus poses awesome therapeutic problems. Table 5 illustrates representative incidences of antibiotic resistance in various countries: The following facts are clearly established:
Table 5: Representative Incidences of Antibiotic Resistance in Various Coun.
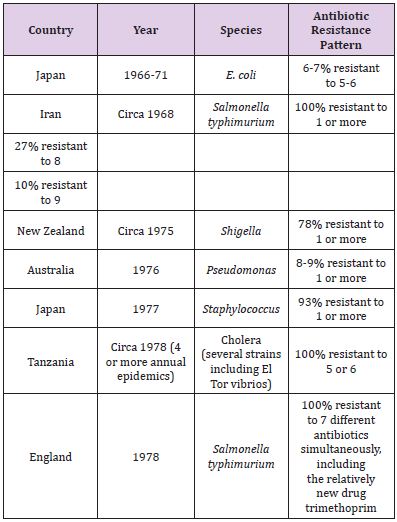
Reference: Adapted and modified from “Germs That Won’t Die” by Marc Lappe, Anchor Press/Doubleday, 1982.
Prior to the use of any antibiotic, a minimal test (called “minimum inhibitory concentration” (MIC) should be undertaken. It consists in taking a culture of the infection, determining the sensitivity of its strains to proposed antibiotics, and assessing that antibiotic and its minimum concentration that will kill the offending pathogen. However, antibiotics are widely inappropriately used as succinctly summarized below:
Nosocomial (or hospital-acquired) infections are ones that are independent of the illness or reason for a patient’s initial hospitalization. Most common nosocomial infections are in rank order: urinary tract infections, pulmonary infections, surgical wound infections, and septic phlebitis. Other patients susceptible to infection are newborn infants, patients with impaired cellular or humoral defense mechanisms, and patients in whom the physical barriers against invasion by pathogens are breached, such as burn or post-surgical patients. They are almost invariably caused by antibiotic-resistant bacteria subsequent to the development of resistant strains of microorganisms within the hospital environment itself. The most common route of infection is the placement of catheters or other devices that pierce the skin of the patient and serve as a natural conduit for infectious organisms.
In most nosocomial infections, the bacteria that take up residence in hospital patients are already resistant to the antibodies currently in use at the same facility. In the case of staph infections, they are also caused by “tolerant” strains that is wherein bacteria withstand the assault of antibacterial agents without actually degrading them or denying them entry into the cell wall. Further, patients who contracted infection(s) in the hospital die in larger percentage than those who acquired their infection in the community. Thus, for patients who have nosocomial infections, 4% of them develop bacteremia (or blood poisoning), the highest risk nosocomial infection.
In the U.S., the statistical records kept by the CDC&P show that typically 3.5% of hospitalized patients acquire nosocomial infections. (In the year 1977, this rate corresponded to 1.5 million patients/year at the then cost in excess of two billion dollars!) This result is obtained despite the fact that we live in an age where reasonable aseptic and hygienic hospital routine is readily attainable with existing technology including surveillance programs in place and inspections by the American Hospital Association/Joint Commission on Accreditation of Hospitals (AHA/ JCAH), without even mentioning means to control antibiotic usage, auditing of records, physician and nurse education, and availability of special consultations.
Antibiotics have become the keystone to mass production of livestock. They seem to make it possible to raise more animals in less space, using less feed, and over a shorter period than through the use of conventional methods. As Table 6 shows, most and in some cases all farm animals receive a substantial portion of antibiotics in their feed at some time during their production:
Table 6: U.S. Use of Antibiotics in Animal Feed.
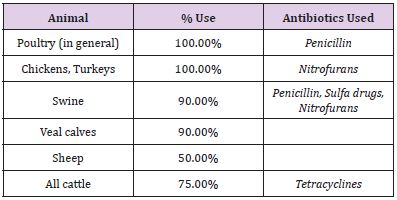
Agricultural applications consume the largest share of the traditional antibiotics market. Nonetheless, the issue is just how much, if at all, does the non-therapeutic use of antibiotics for animals compromise the effectiveness of antibiotics in treating people? This issue has been studied over nearly the past 60 years without any definitive regulatory action:
Notwithstanding the above studies and more recent ones, restraint of the unbridled use of antibiotics supplements in animal feed has not abated in the U.S. By contrast, other nations (Great Britain, former Czechoslovakia, Japan) have recognized that animals can serve as reservoirs of potentially dangerous drugresistant bacteria, and that contamination of one segment of the ecosystem virtually ensures contamination of others.
In 2014, the WHO published its first Global Report on Antimicrobial Resistance with data provided by 114 countries. Antibiotic resistance is no longer a prediction for the future; it is happening right now across the world. This report adds to the 2013 report by the CDC&P, which showed that two million people in the United States are infected annually with antibiotic-resistant bacteria, and 23,000 of them die each year.
Antimicrobial resistance is putting at risk the ability to treat common infections in the community and hospitals. Without urgent, coordinated action, the world is heading towards a postantibiotic era, in which common infections and minor injuries, which have been treatable for decades, can once again kill. As discussed earlier, the development of antimicrobial resistance is a natural phenomenon. However, certain human actions accelerate its emergence and spread. The inappropriate use of antimicrobial drugs, including in animal husbandry, favors the emergence and selection of resistant strains, and poor infection prevention and control practices contribute to further its emergence and spread. Examples of such failures include:
According to new research from the University of East Anglia and Public Health, England, a new and cheap nanopore DNA sequencing technology (named MinION, produced by Oxford Nanopore Technologies, Ltd) on a portable device the size of a USB stick could be used to diagnose infection. The researchers tested the new technology with the complex problem of determining the cause of antibiotic resistance in a new multi-drug resistant strain of the bacterium that causes Typhoid. Current technology for ‘long read’ detailed genomic sequencing can be performed using expensive instrumentation (around £500,000). It is complex to perform, and generally only available in specialist laboratories. The method was demonstrated by successfully mapping the multi-drug resistant genes in a strain Salmonella Typhi haplotype H58, which has recently emerged globally, by pinpointing the exact spot in the chromosomal structure that is home to the genes that make it drugresistant (known as antibiotic resistance island or (ARI)). MinION technology could potentially enable bacterial identification, diagnosis of infectious diseases, and detection of drug-resistance at the point of clinical need.
Precious metals like silver and gold have biomedical properties that have been used for centuries, but how do these materials effectively combat the likes of cancer and bacteria without contaminating the patient and the environment? Gold can be used to either detect or kill cancer cells. Silver is also a very promising material as an antibacterial agent. If you compare silver to current antibiotics, silver does not show drug-resistant behavior.
The atomic structure of nanosilver, revealed by synchrotron X-ray spectroscopy, is proving to be a determinant of silver’s antibacterial activity. When coated with polymers, silver nanoparticles provide a better delivery method for silver to inhibit the bacteria. However, before concluding that nanosilver is an effective and efficient antibacterial agent that could be used to cure human and animal diseases, it must first be determined that it does not attack healthy cells in living systems.
As discussed earlier, taking antibiotics to fight an infection may not be necessary curative. The natural occuring bacteria in the gut harbor several resistance genes and may exchange genes with infectious bacteria, resulting in antibiotic resistance. Therefore, knowing the pool of resistance genes (or “resistome”) in the gut microbiome may improve treatment immensely. Researchers from the Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain), Technical University of Denmark, have just developed a super-fast cheap method (called ‘poreFUME”) that can identify the resistome in 1-2 days. This would enable a treatment of the underlying infection sooner and with better results than traditional approaches. This poreFUME method using nanopore sequencing is very rapid compared to current methods (no culture of the difficult and time-consuming growth of faecal bacteria, device streamed in real-time obviating the need to end a run to access information about the experiment). Today, getting resistome data from a patient takes weeks! In the meantime, the resistome profile might change dramatically further aggravating the patient’s failing health. This approach and methodology may provide better profiling of more patients and hopefully fewer cases of bacterial resistance.
On our individual skin surface and lining our alimentary tract, we are inhabited by trillions of microorganisms some of which are beneficial and protective. But, where do they all come from? The process begins at birth and continues throughout the individual’s lifetime. Their survival can be arrested or their growth inhibited with antibiotics (natural or synthetic). The history of the development of antibiotics was summarized from their discovery in the late 19th century to this day where it continues as a unilateral quest for chemical rather than homeostatic or immunological solutions. In 2016, a novel approach was devised for synthesizing new macrolide antibiotics from simple chemical building blocks. More than 300 new antibiotic candidates have been synthesized, several of which being effective against some of the most stubbornly drug-resistant bacterial strains. This is the first time there is a relatively easy path to synthesize macrolide erythromycin-type antibiotics from scratch.
Antibiotics can be distinguished on the basis of their ability to interfere with the metabolic machinery of microbes. They are supposed to thwart bacterial – not human – cells through one or more of the following modes of action:
Antibiotic activities are clinically separated between those that kill bacteria outright (suicidal or -cidal effect) and those that simply stop the growth of antibiotics (static effect). In practice, however, an antibacterial drug can be bacteriocidal at a high dose and bacteriostatic at a lower one. Its effectiveness can be determined through well-known tests which can be misleading as predictors of clinical efficacy for different antibiotics.
However, the pervasive use of antibiotics may cause the spread of antibiotics’ tolerance and resistance. In a low-level antibiotic assault, or if the antibiotic level remains low or drops, some microorganisms survive, replicate, and form a new “antibioticresistant” infection. Bacteria exist with the innate information needed to resist an antibiotics’ effect even before antibiotics are used and, through an evolutionary mechanism, develop resistance. Evolutionary mechanisms explain both the origins of antibiotic resistance, and the emergence of antibiotic-resistant bacteria. The resistance can spread beyond the initial organisms and simultaneously incorporate resistance to others. “Superbugs” are bacteria resistant to one or more antibiotics. They make it difficult to treat or cure infections that once were easily treated. The bacteria have developed protective genes for resistance and accumulate resistance by developing new genes. Bacteria may have developed resistance to five or six antibiotics so it is not known which antibiotic to use for treatment.
There have been a number of major epidemics due to antibiotic resistance across the world. Antibiotics do not work all the time. If not used judiciously, they develop new strains that are resistant to particular antibiotics. Likewise, a prior course of antibiotic treatment can alter the patient’s susceptibility to a second infection and end up developing new resistant strains. Organisms surviving an initial antibiotic onslaught can pass on their knowledge with dramatic swiftness to another group of organisms, and from there to wider and wider circles of bacterial hosts.
Most common nosocomial (or hospital-acquired) infections are in rank order: urinary tract infections, pulmonary infections, surgical wound infections, and septic phlebitis. They are almost invariably caused by antibiotic-resistant bacteria subsequent to the development of resistant strains of microorganisms within the hospital environment itself.
Antibiotics have also become the keystone to mass production of livestock. Restraint of the unbridled use of antibiotics supplements in animal feed has not abated in the U.S. By contrast, other nations (Great Britain, former Czechoslovakia, Japan) have recognized that animals can serve as reservoirs of potentially dangerous drugresistant bacteria, and that contamination of one segment of the ecosystem virtually ensures contamination of others.
Antibiotic resistance is no longer a prediction for the future; it is happening right now across the world. It is putting at risk the ability to treat common infections in the community and hospitals. Without urgent, coordinated action, the world is heading towards a post-antibiotic era, in which common infections and minor injuries, which have been treatable for decades, can once again kill. The inappropriate use of antimicrobial drugs, including in animal husbandry, favors the emergence and selection of resistant strains, and poor infection prevention and control practices contribute to further its emergence and spread.
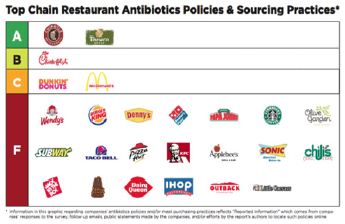
Responding to public pressure, some major food chains and meat suppliers have pledged to use fewer antibiotics. A report contributed to by the environmental nonprofit National Resources Defense Council and several other public groups analyzed the practices and policies of 25 of the largest fast food and fast-casual restaurants in the U.S. to see how companies were faring. Each chain was given a letter grade based on its use of antibiotics and its transparency about it. The report card is shown in Figure 4.
Nanomaterials (particularly nanogold and nanosilver) are becoming effective and efficient ant-bacterial agent for curing human (and animal) diseases. Nanopore technology is bringing us closer to a personalized antibiotic treatment, particularly in cases of drug resistance.


