Abstract
We developed a rapid and practical screening method for simultaneously detecting both Escherichia coli and bacteria harboring the mcr-1 colistin resistance gene using high-speed real-time polymerase chain reaction with specific TaqMan probes. The entire procedure, from sample processing to the final result, was performed within 1 h. The practical utility of this method was verified by analyzing 27 retail meat samples for the presence of colistin-resistant bacteria. The results indicated the potential of this method for the convenient and rapid screening of food items to detect contamination with mcr-1–positive bacteria, which can be especially useful for on-site testing in developing countries.
Keywords: Colistin-Resistant Bacteria; Food; Mcr-1; Rapid Screening Method; Real- Time Polymerase Chain Reaction
Introduction
Colistin is recognized as one of the few remaining available antibiotics for the treatment of intractable infections caused by multidrug-resistant Gram-negative bacteria [1]. Recent studies have shown that bacteria carrying the mcr gene, which confers colistin resistance to most members of the Enterobacteriaceae, are widely disseminated, particularly in Asia [2,3]. Since colistin is widely used in animal husbandry [4], the spread of colistin-resistant (CR) bacteria in communities via livestock food is a potential risk factor. Moreover, CR bacteria are often found in animals and animalfood [5-7]; thus, monitoring CR bacteria in animal-food is essential. However, the conventional culture method [8] for detecting CR bacteria in food is laborious and time-consuming. Rapid detection of colistin resistance genes at the research level is now possible using the SYBR green method [9], but its widespread practicality is limited due to the need for complex steps and equipment involved in DNA extraction from samples and determination of result specificity. To overcome this limitation, we here report a simple, rapid, and practical detection method of Escherichia coli harboring mcr-1, as a representative CR bacterium, using a highspeed real-time polymerase chain reaction (PCR) kit. We further verified the utility of this method for detecting CR bacteria in retail meat samples. Although a real-time PCR assay for the detection of mcr genes from bacterial isolates has already been established, this newly proposed detection method holds practical relevance for widespread use, as the entire procedure, from food sample processing to the final result, can be completed within only 1 h.
Materials and Methods
A total of 27 retail meat samples, including pork and chicken,
were collected from 10 markets (two supermarkets and eight
local traditional markets) in Vietnam and five supermarkets in
Japan during November and December 2019. None of the eight
traditional markets in Vietnam maintained a refrigerator for meat
preservation. In contrast, the two supermarkets in Vietnam and
all five supermarkets in Japan had refrigerators for food storage.
Each sample was collected from one meat type per market.
Bacterial cultures and DNA extraction were performed on the
collection day. Ten grams of each meat sample were placed in a
stomacher bag (AS ONE, Osaka, Japan) containing 90 mL buffered
peptone water. The samples were hand-homogenized for 2
min. The resulting homogenate was inoculated on CHROMagar
COL-APSE (CHROMagar, Paris, France), a selective medium for
CR Gram-negative bacteria, and cultured at 37 °C for 24 h. CR E.
coli-like colonies were distinguished based on colony color (dark
pink to reddish) after cultivation [8,10]. A representative colony
was isolated by sub-culturing on MacConkey agar, and bacterial
identification was performed. The colistin minimum inhibitory
concentration (MIC) was estimated, and colistin resistance
genes (mcr-1 to -5) were detected by multiplex PCR as described
previously [6,11].
In parallel, DNA was extracted from 1 mL of the homogenate
using the Kaneka Easy DNA Extraction Kit version 2 (Kaneka,
Tokyo, Japan). The presence of E. coli and the colistin resistance
gene mcr-1 in the DNA extracts was determined by real-time PCR
using a mobile PCR device, PicoGene PCR1100 (Nippon Sheet
Glass, Tokyo, Japan). PCR primers and TaqMan probes for realtime
PCR detection of E. coli 16S rRNA and mcr-1 were prepared
as described previously (Table 1) [12]. Details regarding the realtime
PCR, including PCR mixtures and thermal cycling conditions,
are provided in Tables 2 & 3, respectively. The DNA extract of the
CR E. coli strain (E362) [6] carrying mcr-1 was utilized as a positive
control in PCR. The entire 50 PCR cycles were completed within only
21 min. Moreover, the real-time PCR device could simultaneously
measure fluorescence at three different wavelengths for the same
sample. Two fluorescent dye-labeled TaqMan probes (Integrated
DNA Technologies, Singapore), Cy5 for E. coli 16S rRNA and FAM for
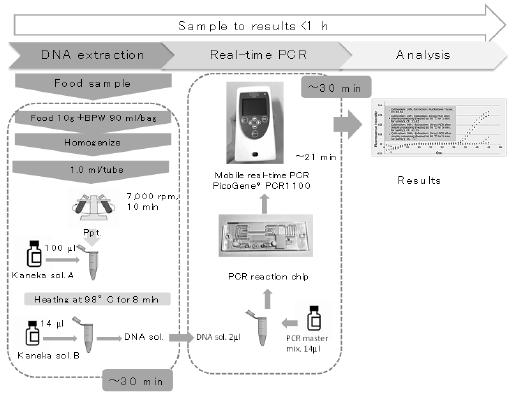
mcr-1, were used for each sample. The entire protocol is outlined
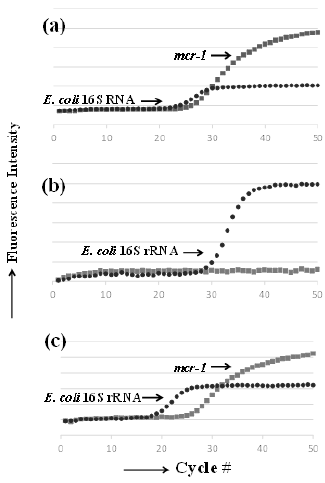
in Figure 1. Figure 2 shows representative real-time PCR profiles
of the samples.
Figure 1: Outline of the screening protocol using mobile real-time PCR PicoGene® PCR1100. BPW, buffered peptone water.
Figure 2: Representative plots obtained from real-time
PCR amplification of mcr-1 and E. coli 16S rRNA genes in
meat samples.
a) Positive control, mcr-1 E. coli.
b) mcr-1–negative pork sample, H-A market pork.
c) mcr-1–positive chicken sample, H-E market chicken.
Results and Discussion
The detection sensitivity of the method was assessed using pork meat samples spiked with an mcr-1-positive E. coli strain culture. The lower limit of mcr-1-E. coli detection for the entire method, from DNA extraction to detection by real-time PCR, was 7 × 102 CFU/g; however, a minimum of 7 × 103 CFU/g was required for quantification using a linear correlation. In the validation study using retail meat samples, CR E. coli-like bacteria were detected using the culture-based method in eight out of ten chicken and in three out of seven pork samples purchased in Vietnam (Table 4). The semi-quantitative levels of CR bacteria in these samples were in the range 103‒108 CFU/g (Table 4). All representative CR E. coli isolates from each sample were confirmed to be resistant to colistin (MIC ≥ 4 μg/mL) and possessed mcr-1 but not mcr-2 to -5, except for the H-E market pork sample, which harbored mcr-3 in addition to mcr-1, as determined by multiplex PCR. No samples from the Japanese supermarkets were contaminated with CR bacteria. All samples, except for the H-E market pork sample, that were positive via the culture-based method were also positive by real-time PCR (Table 4). Some culture-negative samples such as H-B market pork, T-B market chicken, T-B market pork, and T-E market chicken were PCR-positive. Such contradictory results may be attributed to the features of the real-time PCR method and its ability to detect mcr-1 even in dead cells and/or non-E. coli cells. In contrast, a pork sample from the H-E market showed CR E. coli colonies after culturing but tested negative for mcr-1 by real-time PCR. Such discrepant cases could be due to a low level of mcr-1–positive bacteria below the detection limit of the real-time PCR method or the presence of bacteria expressing non-mcr CR determinants [13].
The new method presented herein detects the target gene and facilitates quantitative analysis. In addition, the method using TaqMan probes has high detection specificity, and is simple because it does not require specificity verification by melting curve analysis, even for one-step extracted DNA from food. The results output the ratio of bacteria carrying mcr-1 to the total number of E. coli cells, which may be mcr-1–positive or –negative bacteria (Figure 2). The detected quantitative mcr-1 levels were higher than the CR E. coli-like bacterial levels determined via the culture-based method because the real-time PCR method detects all mcr-1 regardless of bacterial species. The quantitative linear range detected via realtime PCR was between 103 and 106 CFU/g. Although the detected signal was below the quantitative linear range limit in some samples, they were still considered to have positive results via realtime PCR. The approach described in this study provides limited information regarding the degree of contamination; nevertheless, the developed method is reliable and practical owing to a short processing time, enabling the rapid screening of contaminating bacteria with mcr-1 in food.
Conclusion
A new rapid and practical screening method was developed for detecting CR E. coli in food samples. The developed method is advantageous because it is easy to perform, has a short processing time, and provides reliable results that are consistent with those obtained by traditional methods.
Declarations
Ethics Approval and Informed Consent
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
This study was conducted using a mobile PCR device, PicoGene PCR1100, provided by Nippon Sheet Glass, Tokyo, Japan. There were no competing financial interests or personal relationships that could have influenced the work reported in this paper.
Funding
This work was supported by the Japan Society for the Promotion of Science KAKENHI (grant number 20H00561).
Authors’ Contributions
HV participated in the study design, sample collection, and real-time PCR assay, and drafted the manuscript. CA-K performed the real-time PCR assay. KT participated in the study design. RK performed PCR assays and microbial analysis. DTK, TNN, HTT, CDV, and PDN contributed to sample collection and bacterial cultures. YY contributed to the study design, supervised data collection, and drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgment
Not applicable.
Availability of Data and Materials
The datasets used and/or analyzed in this study are available from the corresponding author on reasonable request.
References
- Giamarellou H, Poulakou G (2009) Multidrug-resistant gram-negative infections: what are the treatment options? Drugs 69(14): 1879-1901.
- Shen Y, Zhou H, Xu J, Wang Y, Zhang Q, et al. (2018) Anthropogenic and environmental factors associated with a high incidence of mcr-1 carriage in humans across China. Nat Microbiol 3(9): 1054-1062.
- Yamamoto Y, Kawahara R, Fujiya Y, Sasaki T, Hirai I, et al. (2018) Wide dissemination of colistin-resistant Escherichia coli with the mobile resistance gene mcr in healthy residents in Vietnam. J Antimicrob Chemother 74(2): 523-524.
- Rhouma M, Beaudry F, Letellier A (2016) Resistance to colistin: what is the fate for this antibiotic in pig production? Int J Antimicrob Agents 48(2): 119-126.
- Kawahara R, Fujiya Y, Yamaguchi T, Khong DT, Nguyen TN, et al. (2019) Most domestic livestock possess colistin-resistant commensal Escherichia coli harboring mcr in a rural community in Vietnam. Antimicrob Agents Chemother 63(6): e00594-19.
- Yamaguchi T, Kawahara R, Harada K, Teruya S, Nakayama T, et al. (2018) The presence of colistin resistance genes mcr-1 and -3 in ESBL-producing Escherichia coli isolated from food in Ho Chi Minh City, Vietnam. FEMS Microbiol Lett 365(11): fny100.
- Perez-Rodriguez F, Mercanoglu Taban B (2019) A state-of-art review on multi-drug resistant pathogens in foods of animal origin: risk factors and mitigation strategies. Front Microbiol 10: 2091.
- Abdul Momin MHF, Bean DC, Hendriksen RS, Haenni M, Phee LM, et al. (2017) CHROMagar COL-APSE: a selective bacterial culture medium for the isolation and differentiation of colistin-resistant Gram-negative pathogens. J Med Microbiol 66(11): 1554-1561.
- Tolosi R, Apostolakos I, Laconi A, Carraro L, Grilli G, et al. (2020) Rapid detection and quantification of plasmid-mediated colistin resistance genes (mcr-1 to mcr-5) by real-time PCR in bacterial and environmental samples. J Appl Microbiol 129(6): 1523-1529.
- Thiry D, Berrah A, Evrard J, Duprez JN, Mainil JG, et al. (2019) Assessment of two selective agar media to isolate colistin-resistant bovine Escherichia coli: Correlation with minimal inhibitory concentration and presence of mcr genes. J Microbiol Methods 159: 174-178.
- Wakabayashi Y, Sekizuka T, Yamaguchi T, Fukuda A, Suzuki M, et al. (2020) Isolation and plasmid characterisation of Salmonella enterica serovar Albany harboring mcr-5 from retail chicken meat in Japan. FEMS Microbiol Lett 367(15): fnaa127.
- Daniels JB, Campbell D, Boyd S, Ansari U, Lutgring J, et al. (2019) Development and validation of a clinical laboratory improvement amendments-compliant multiplex real-time PCR assay for detection of mcr Microb Drug Resist 25(7): 991-996.
- Aghapour Z, Gholizadeh P, Ganbarov K, Bialvaei AZ, Mahmood SS, et al. (2019) Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect Drug Resist 12: 965-975.

 Short Communication
Short Communication