ABSTRACT
The objective of the present study was to conduct DNA-based genotyping assay, specifically exploring microsatellite regions, on wheat M2 population for identifying the genuine mutant plants (other than the natural variants) before conducting TILLING experiments. Seed of a wheat variety ‘NN-Gandum-1’ was exposed to two different time exposure and temperatures i.e., 1 and 2 hours and 33°C and 35°C, respectively. It was established that 0.8% EMS when exposed for 2 hours at 35°C was found appropriate for developing a suitable TILLING population. For determining the quality of the mutant population, a purity analysis was carried out using SSR primer pairs with high PIC values. Initially, 77 SSRs were surveyed on wheat genotypes. In total, 20 SSRs showing high PIC values were selected for surveying 3634 M2 plants. Out of these, three SSR primer pairs WMS-46, WMS-249 and WMS-311 detected natural variants in M2 TILLING population with 5.09, 7.23 and 8.29%, respectively. The While, WMS-249 and WMS-311 were located in densely mapped regions (near the centromere) than that of the WMS-46. It was concluded that 20 SSRs with high PIC value are sufficient to identify true-to-type mutants (other than natural variants) in a wheat TILLING population. This study has implications for identifying genes and their function as well as in genetic improvement of wheat. This information can also be translated on other crops.
Keywords: Triticum Aestivum; Tilling Population; Natural Variants; Simple Sequence Repeat (SSR); Genomics- Breeding
Abbreviations: SSR: Simple Sequence Repeat; NIBGE: National Institute for Biotechnology & Genetic Engineering; TILLING: Targeting Induced Local Lesions IN Genomes; EMS: Ethyl Methane Sulphonate; CTAB: Cetyl Trimethyl Ammonium Bromide; IAEA: International Atomic Energy Agency; CRD: Complete Randomized Design; IWGSC: International Wheat Genome Sequencing Consortium
Introduction
Wheat provides 20% of the total calories consumed by human
beings. Major cultivated species of wheat is Triticum aestivum L.
(2n=6x=42, also known as bread wheat). It has three sub genomes
A, B and D, together makes 17.6 GB wheat genome Spannagl, et
al. [1]. In spite of huge investment made in understanding wheat
Genet. and genomics, there are still many gaps in elucidating different genetic pathways because of its huge and complex genome
size and its ploidy level Slade, et al. [2]. TILLING (Targeting Induced
Local Lesions IN Genomes), a powerful reverse genetic strategy
that allows the detection of induced point mutations in individuals
of the mutagenized populations (to induce genetic variations, a
number of mutagens like physical and chemical have been used. Out
of which, chemical mutagen especially ethyl methane sulphonates
(EMS)-induces point mutations randomly in genomes of different
crop species Greene, et al. [3-6], can address the major challenge
of linking sequence information to the Biol. function of genes and
can also identify novel variation for crop Breeding. Slade, et al. [2].
Wheat is especially well- suited for TILLING due to the high
mutation densities tolerated by polypoid. However, only a few
wheat TILLING populations are currently available in the world,
which is far from satisfying the requirement of Res.ers Chen, et
al. [7-13,5]. Before inducing mutations, genetic uniformity of the
genetic material is extremely important Sabetta, et al. [14]. Seed
purity is undertaken by morphological means. Due to uncertainties
in such methods, there is always a risk of incorrectly rejecting or
accepting a seed lot Remund, et al. [15]. Morphological traits are
growth stage specific, prone to environmental errors and are
limited in number. Wheat is a self-pollinating plant but occasionally
outcrossing occurs. However, it is impractical to have pollen control
by bagging spikes in the greenhouse or in the field. Therefore, rapid
and accurate assessment of a large TILLING population through
such means is difficult. Thus, as a standard procedure, putative
mutants identified are further subjected to microsatellite marker
analysis for confirming their authenticity Wu, et al. [16-20]. Simple
Sequence Repeats (SSRs) are co-dominant in expression-make them
a marker of choice and have been used extensively in genotyping
individuals of different plant species Asif, et al. [21].
The propensity of SSRs showing hyper variability has been
exploited to study the natural polymorphisms found in TILLING
population of different crop species Xin, et al. [22,14] and or
identifying contamination (off type plants) in mutant population
that usually occurs by several means including mixing at harvesting,
threshing, etc. Bora, et al. [19]. We consider this approach as a costeffective
way to maintain quality control of the mutant stock. Thus,
it is extremely important to test for heterogeneity and or natural
variants in TILLING population.
Material and Methods
Plant Material
The hexaploid NN-Gandum-1 (Triticum aestivum L., 2n=6X=42) is a spring wheat variety released for cultivation in July 2016 that was bred by making a cross (Chirya-3/Opata//2x parula/3/ Rohtas-90) at the Plant Genomics & Mol. Breeding Lab, National Institute for Biotechnol. and Genetic Engineering. NN-Gandum-1 (also called as Gandum-1)’ has been evaluated for studying its adaptability throughout the Punjab province in Pakistan.
Mutagenesis and Tilling Population Development
Genetic purity of the variety was ensured by planting 500 single spike rows. Out of these, only one line was selected by observing uniformity in different morphological traits. Then the seed was increased by planting small plot. This seed was used to expose to chemical mutagen EMS. For optimization of dose, 180 seed in three replicates of each line were treated with eight different EMS doses (0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1%), for two different exposure times (16 h and 18 h) at room temperature (RT = 25°C) with continuous shaking.
DNA Isolation
Genomic DNA was isolated from leaves of 21-day-old seedlings of Gandum-1 and each of 74 the M2 plants using the modified cetyl trimethyl ammonium bromide (CTAB) method. The DNA quantification was carried out using Nano-dropTM1000 spectrophotometer (Thermo scientific, USA) and diluted accordingly. The quality and quantity of the genomic DNA was also determined on 0.8% agarose gel. SSR purity analysis on M2: A total of 77 SSR primer pairs (WMS series) exhibited high PIC values were selected by surveying on 96 wheat genotypes (unpublished data). Total volume of 20 μL was used for PCR. The ingredients of the PCR were 20 ηg genomic DNA, 0.2mM dNTP, 1.5mM MgCl2, 2 μg of each primer (forward and reverse) and 0.5U Taq polymerase. The SSR (PCR) amplification of genomic DNA was carried out by incubating the DNA samples at 95°C for 5 min, then 35 cycles comprising 94°C for 1 min, annealing of primer at 55- 60°C for 30 sec and then extension at 72°C for 1 min. The final extension was carried out at 72°C for 10 min in Thermal Cycler (Bio-RAD C1000 Touch). After PCR amplifications, the PCR products were separated on 2.4 % high-resolution agarose gel (Bio-RAD electrophoresis system). The gels were stained with ethidium bromide and photographed on GelDoc-It®2 310 Imager.
Chromosome Survey and Statistical Analysis
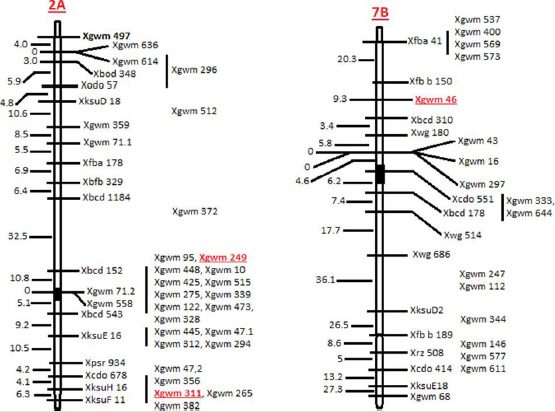
Position of the SSRs on chromosome was determined using Ensembl Plants (http://archive.plants.ensembl.org/index.html). Physical map of chromosome, position of markers and position of centromere was collected from the International Wheat Genome Sequencing Consortium (IWGSC) website (https://www. wheatgenome.org/) (Figure 1). For the analysis of EMS experiment, complete randomized design (CRD) was used. Analysis of variance (ANOVA) of the data was conducted using statistix 8.1 software.
Results
In present study, seed of wheat variety Gandum-1 were
exposed to EMS at two exposure times (1 and 2 h) as previously
reported Bahar, et al. [23]. It was observed that the germination
rate showed gradual depression with the gradual increment of
EMS concentration. After exposing for 1 hours, the germination
percentage at 0.4, 0.5, 0.6, 0.7, 0.8 and 0.9% (v/v) EMS
concentration was 83.4, 72.5, 58.3, 47.6, 42.2 and 34.5 respectively.
While after exposing for 2 hours, the germination percentage at
0.4, 0.5, 0.6, 0.7, 0.8 and 0.9 % EMS concentration was 79.3, 67.8,
53.4, 41.2, 36.7, and 29.5, respectively. From this experiment, 0.8%
EMS concentration for 2 hours exposure time at 35°C was found
appropriate for developing a wheat TILLING population. Before
conducting TILLING experiments, purity analysis of the newly
developed genetic resource (M2) was done for differentiating
induced mutations from the natural once. The M1 plants that
were raised from EMS-mutagenized seed, were self-fertilized for
harvesting M2 seed. In next wheat growing season, M2 seed were
sown to raise M2 plants. From each of the M2 plant (in total 3634),
genomic DNA was extracted. Genomic DNA of each of the plant was
normalized up to 20ng/ul. PCR conditions were optimized.
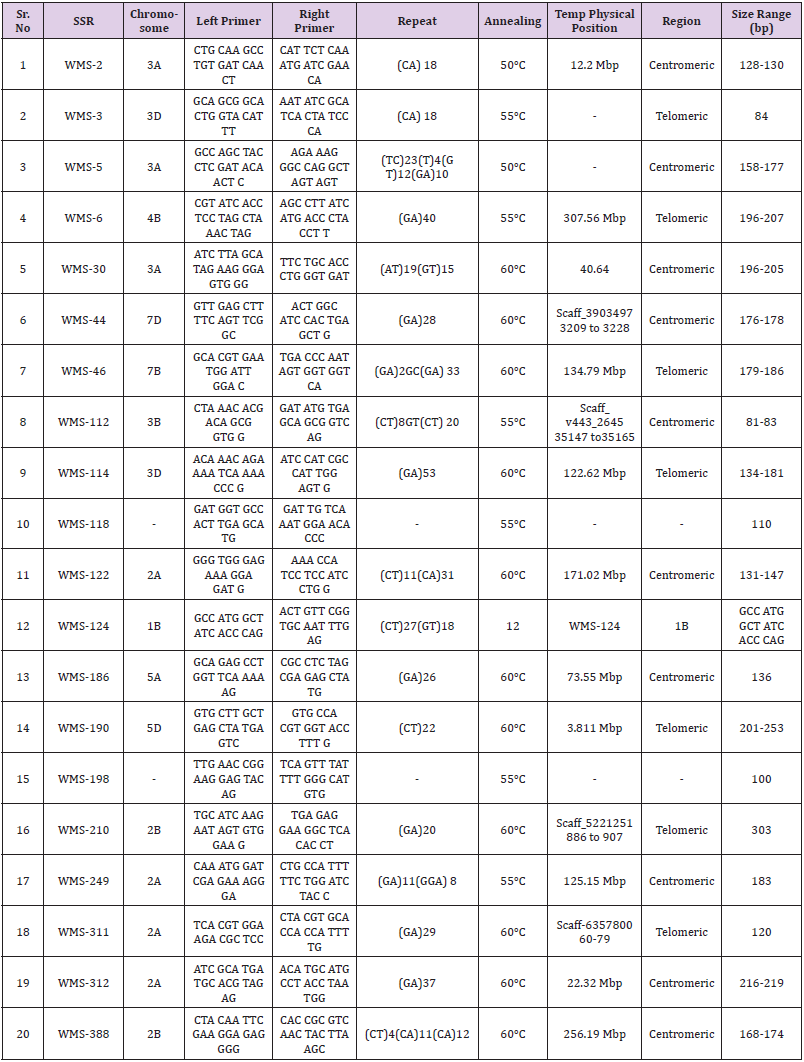
In total, 77 SSRs were surveyed on 96 wheat genotypes (data
unpublished). Out of these, 20 SSRs primer pairs (WMS-2, 107 WMS-
3, WMS-5, WMS-6, WMS-30, WMS-44, WMS-46, WMS-112, WMS-
114, WMS-118, WMS- 122, 108 WMS-124, WMS-186, WMS-190,
WMS-198, WMS-210, WMS-249, WMS-311, WMS-312 and WMS-
388) (Table 1) were selected on the basis of their PIC values. From
these 20 primer pairs, only three SSR primer pairs (WMS-46, WMS-
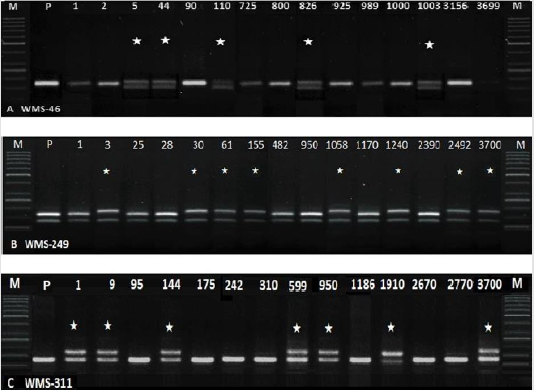
249 and WMS-311) amplified polymorphic amplicons in M2 plants
(Figure 2). These plants appeared to contain natural mutations.
Two SSRs were mapped on chromosome 2A (WMS-249 and WMS-
311) while one was mapped on chromosome 7B (WMS-46) (Figure
1). The frequency of natural variants detected by WMS-46, WMS-
249 and WMS-311 were 5.09% (188 out of 3634), 7.23% (in total
268) and 8.29% (in total 307), respectively. These variants showing
polymorphic alleles other than wild type (amplified by either of the
primer pairs) were excluded from the TILLING population to avoid
natural contamination (as per requirement of TILLING).
Figure 2: Amplification of M2 samples with WMS-46, 249 and 311. Lane M=50bp Ladder (P= parent, NN Gandum-1. *=natural variants).
Discussion
Dose of mutagen is largely dependent upon the genetic makeup of a genotype. It has been proposed by some Researchers that large genomes necessitate higher EMS concentrations. For example, reduction in germination rate in wheat is an indication of effective mutagenesis Bahar, et al. [23]. Such reduction in or slowing down of wheat seed germination with enhanced EMS concentration may occur by the delay or inhibition of some physiological processes Kumar, et al. [24-26]. An ideal population would carry maximum mutations without losing fertility of majority of plants Weil, et al. [27]. Thus, before launching an experiment, dose of a mutagen should be optimized Till, et al., [28-29]. Induction of mutations in crop species seems to be a straightforward strategy but some technical challenges including purity of a genetic material impact the quality of a newly developed mutant population Caldwell, et al., [30]. Determining the genetic purity of a mutant population is one of the most important features that aids in improving the quality of a TILLING experiments by discarding the natural variants present in the population. Microsatellite markers have also been used for purity analysis of TILLING populations of fruit, vegetables, sunflower, Soybean and other crops Wu, et al. [16,22,14,31]. For example, a total of 4 and 15 SSR primer pairs were surveyed on sunflower and sorghum, respectively to identify variants in population Xin, et al. [22,14].
Conclusion
It is concluded that SSR analysis by selecting the highly polymorphic loci (with high PIC value), preferably 20 SSRs, can be undertaken for selecting true-to-type mutant plants (other than natural variants). Usually, one can screen the whole population (3634 or more plants) within two week using semiautomated genotyping system. Therefore, the survey of SSR loci for determining the purity of a mutant population is recommended before embarking TILLING strategy, exome capturing Henry, et al. [32-34], SNP discovery Rimbert, et al. [35], re-sequencing of mutants of TILLING population Kagale, et al. [36] and genomebased strategies Scossa, et al. [37-39] that would help in reaching gold standard conclusions.
Acknowledgment
This work is supported by the International Atomic Energy Agency (IAEA), Vienna Austria through a project entitled “Development of novel germplasm through TILLING in crop plants using mutation and genomic approaches (Pak/5/047)”.” Any opinions, findings, conclusions, or recommendations expressed in this manuscript are those of the author(s) and do not necessarily reflect the views of the International Atomic Energy Agency (IAEA).
References
- Spannagl M, T Nussbaumer, K C Bader, M M Martis, M Seidel, et al. (2016) Pgsb plantsdb: Updates to the database framework for comparative plant genome research. Nucleic acids research 44: D1141-D1147.
- Slade A J, S I Fuerstenberg, D Loeffler, M N Steine, D Facciotti (2005) A reverse genetic, nontransgenic approach to wheat crop improvement by tilling. Nature biotechnology 23: 75-81.
- Greene E A, C A Codomo, N E Taylor, J G Henikoff, B J Till, et al. (2003) Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in arabidopsis. Genetics 164(2): 731-740.
- Kodym A, R Afza (2003) Physical and chemical mutagenesis. In: Plant functional genomics Springer, pp. 189-203.
- Hussain M, M A Iqbal, B J Till, Rahman M (2018) Identification of induced mutations in hexaploid wheat genome using exome capture assay. PLOS ONE 13(8): e0201918.
- Hussain M, M Rahman (2019) Registration of pgmb-15-30 spring wheat. Journal of Plant Registrations 13(2).
- Chen L, L Huang, D Min, A Phillips, S Wang, et al. (2012) Development and characterization of a new tilling population of common bread wheat (triticum aestivum l). PLoS One 7: e41570.
- Rawat N, S K Sehgal, A Joshi, N Rothe, D L Wilson, et al. (2012) A diploid wheat tilling resource for wheat functional genomics. BMC Plant Biology 12: 1.
- Uauy C, K Krasileva, P Bailey, V Buffalo, A Phillips, et al. (2013) An in-silico functional genomics resource: Targeted re-sequencing of wheat tilling mutant populations. In: Plant and Animal Genome XXI Conference.
- Tadele Z (2016) Mutagenesis and tilling to dissect gene function in plants. Current Genomics 17(6): 499-508.
- Mo Y, T Howell, H Vasquez-Gross, L A de Haro, J Dubcovsky, et al. (2017) Mapping causal mutations by exome sequencing in a wheat tilling population: A tall mutant case study. Molecular Genetics and Genomics, p. 1-15.
- Szurman-Zubrzycka M E, J Zbieszczyk, M Marzec, J Jelonek, B Chmielewska, et al. (2018) Hortillus-a rich and renewable source of induced mutations for forward/reverse genetics and pre-breeding programs in barley (hordeum vulgare l.). Frontiers in Plant Science 9: 216.
- Till BJ, S Datta, J Jankowicz Cieslak (2018) Tilling: The next generation. Adv Biochem Eng Biotechnol 164: 139-160
- Sabetta W, V Alba, A Blanco, C Montemurro (2011) Suntill: A tilling resource for gene function analysis in sunflower. Plant Methods 7(1): 20.
- Remund K M, D A Dixon, D L Wright, L R Holden (2001) Statistical considerations in seed purity testing for transgenic traits. Seed Science Research 11(2): 101-120.
- Wu J L, C Wu, C Lei, M Baraoidan, A Bordeos, et al. (2005) Chemical-and irradiation-induced mutants of indica rice ir64 for forward and reverse genetics. Plant molecular biology 59(1): 85-97.
- Wang L, L Chang, H Li, L Ge, A Xin, et al. (2009) Method of testing wheat seeds purity by molecular markers. J Triticeae Crops 29: 1-8.
- Wang L, L Liu, F Zhang, H Li, B Pang, et al. (2014) Detecting seed purity of wheat varieties using microsatellite markers based on eliminating the influence of non-homozygous loci. Seed Science and Technology 42(3): 393-413.
- Bora A, P R Choudhury, V Pande, AB Mandal (2016) Assessment of genetic purity in rice (oryza sativa l.) hybrids using microsatellite markers. 3 Biotechnology 6: 1-7.
- Sun H, S Wang, Y Dong, C Zhang, B Peng, et al. (2018) Application of ssr markers for purity testing of commercial hybrid soybean (glycine max l.).
- Asif M, M Rahman, J I Mirza, Y Zafar (2009) Parentage confirmation of cotton hybrids using molecular markers. Pakistan Journal of Botany 41(2): 695-701.
- Xin Z, ML Wang, NA Barkley, G Burow, C Franks, et al. (2008) Applying genotyping (tilling) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biology 8(1): 103.
- Bahar B, M S Akkaya (2009) Effects of ems treatment on the seed germination in wheat. Journal of applied biological sciences 3(1): 53-58.
- Kumar G, P Gupta (2009) Induced karyomorphological variations in three phenodeviants of capsicum annuum l. Turkish Journal of Biology 33(2): 123-128.
- Devi A S, L Mullainathan (2011) Physical and chemical mutagenesis for improvement of chilli (capsicum annuum L). World Appl Sci J 15(1): 108-113.
- Borovsky Y, Y Tadmor, E Bar, A Meir, E Lewinsohn, et al. (2013) Induced mutation in β-carotene hydroxylase results in accumulation of β-carotene and conversion of red to orange color in pepper fruit. Theoretical and applied genetics 126(3): 557-565.
- Weil CF, R A Monde (2007) Getting the point-mutations in maize. Crop science 47: S-60- S-67.
- Till BJ, J Cooper, TH Tai, P Colowit, EA Greene, et al. (2007) Discovery of chemically induced mutations in rice by tilling. BMC plant biology 7: 1.
- Hussain M, M Gul, R Kamal, M A Iqbal, S Zulfiqar, et al. (2021) Prospects of developing novel genetic resources by chemical and physical mutagenesis to enlarge the genetic window in bread wheat varieties. Agriculture 11(7): 621.
- Caldwell D G, N McCallum, P Shaw, G J Muehlbauer, D F Marshall, et al. (2004) A structured mutant population for forward and reverse genetics in barley (hordeum vulgare l). The Plant Journal 40(1): 143-150.
- Mazzucato A, F Cellini, M Bouzayen, M Zouine, A Petrozza, et al. (2015) A tilling allele of the tomato aux/iaa9 gene offers new insights into fruit set mechanisms and perspectives for breeding seedless tomatoes. Molecular Breeding 35(22): 1-15.
- Henry I M, U Nagalakshmi, M C Lieberman, K J Ngo, K V Krasileva, et al. (2014) Efficient genome-wide detection and cataloging of ems-induced mutations using exome capture and next-generation sequencing. The Plant Cell 26(4): 1382-1397.
- Krasileva KV, H A Vasquez-Gross, T Howell, P Bailey, F Paraiso, et al. (2017) Uncovering hidden variation in polyploid wheat. Proceedings of the National Academy of Sciences 114(6): 201619268.
- Uauy C, BB Wulff, J Dubcovsky (2017) Combining traditional mutagenesis with new high-throughput sequencing and genome editing to reveal hidden variation in polyploid wheat. Annual Review of Genetics 51: 435-454.
- Rimbert H, B Darrier, J Navarro, J Kitt, F Choulet, et al. (2018) High throughput snp discovery and genotyping in hexaploid wheat. PloS one 13(1): e0186329.
- Kagale S (2016) Tilling by sequencing for genome-wide mutation discovery and functional genomics in camelina sativa. In: Plant and Animal Genome XXIV Conference. Plant and Animal Genome.
- Scossa F, Y Brotman, F d A e Lima, L Willmitzer, Z Nikoloski, et al. (2016) Genomics-based strategies for the use of natural variation in the improvement of crop metabolism. Plant Science 242: 47-64.
- Burkart Waco D, H Tsai, K Ngo, I M Henry, L Comai, et al. (2017) Next-generation sequencing for targeted discovery of rare mutations in rice. In: Biotechnologies for plant mutation breeding Springer, pp. 323-340.
- Torkamaneh D, B Boyle, F Belzile (2018) Efficient genome-wide genotyping strategies and data integration in crop plants. Theoretical and Applied Genetics: 131: 499-511.

 Research Article
Research Article