Abstract
Chronic myelomonocytic leukemia is a clonal hematopoietic stem cell disorder with overlapping features of myelodysplastic syndromes and myeloproliferative neoplasms. Median age at diagnosis is 70 years and, in many cases, it is diagnosed occasionally. Bone marrow karyotype is normal in two thirds of patients, with a few recurrent aberrations including -Y, -7, and +8. Here, we report the case of a patient affected by dysplastic CMML-1 subtype according to the 2017 WHO classification, showing a 46,XX,del(7) (q21q36),t(4;12)(q24;q15)[18]/46,XX[2] peculiar karyotype. To assess the real nature of these chromosomal abnormalities we performed a Cancer CGH+SNP array. On chromosome 7q, we identified three noncontiguous deletions at bands q21.11-q22.1, q22.1-q32.2 and q34-q36.1, while we did not detect any copy number neutral loss. In addition, the SNP array unveiled the unbalanced nature of t(4;12), with three cryptic genomic imbalances: two deletions on chromosome 4, at bands q13.1-q13.3 and q24, and one deletion on chromosome 12, at bands q21.33-q23.1. These three deletions are known to involve many OMIM genes, including TET2 (OMIM *612839) and NFKB1 (OMIM *16401). Chromosome 7 aberrations, detected in about 20% of CMML patients with cytogenetic abnormalities, have been recognized as an adverse prognostic factor, therefore allocating to the high cytogenetic risk category. Several tumor suppressor genes map in the chromosome 7 deleted regions, such as EZH2, SAMD9L and CUX1. Deletion of these regions can contribute to disease progression and could account for the differences in patients’ prognosis due to the variability of breakpoint regions on 7q. Next-generation sequencing (NGS) analysis confirmed this result revealing a double TET2 mutation. Therefore, we underline the role of CGH arrays in CMML diagnostic workup. These tools, together with NGS, represent a valid instrument to provide insight not only in molecular pathogenesis but also in disease progression.
Keywords: Chronic Myelomonocytic Leukemia; aCGH; Chromosome 7q; Cryptic Genomic Imbalances; NGS
Abbreviations: CMML: Chronic Myelomonocytic Leukemia; MDS: Myelodysplastic Syndromes; BM: Bone Marrow; NGS: Next-Generation Sequencing
Case Report
Chronic myelomonocytic leukemia (CMML) is a clonal hematopoietic stem cell disorder with overlapping features between myelodysplastic syndromes (MDS) and myeloproliferative neoplasms and an inherent leukemic risk of ~15% over 3-5 years [1,2]. The 2017 WHO classification has recommended its partitioning into three categories based on peripheral blood and bone marrow (BM) blasts percentage [2]. In addition, the previously used 1994 FAB Cooperative Leukemia Group subdivision into a “dysplastic” (MD) and a “proliferative” CMML variant has been revived. Median age at diagnosis is 70 years, with a male preponderance. In many cases the diagnosis is occasional, with a median survival of 24-36 months [3]. Over the years several studies aimed to identify clinical and biological features associated with CMML survival outcomes, leading to the development of different prognostic models for individual patients’ treatment decision-making [4]. Like acute myeloid leukemia, CMML patients demonstrate ~10-15 mutations per kilobase of coding DNA regions, [5] while clonal cytogenetic abnormalities are observed in 20-30% of cases, including +8, -Y, chromosome 7 abnormalities, +21, and complex karyotypes [1]. In 2014 an international collaborative study between Mayo clinic and French consortium stratified CMML patients into three cytogenetic risk groups: high: complex karyotype, chromosome 7 abnormalities, monosomal karyotype; intermediate: +8, +21, others; and low: normal karyotype, -Y, der(3q) [3].
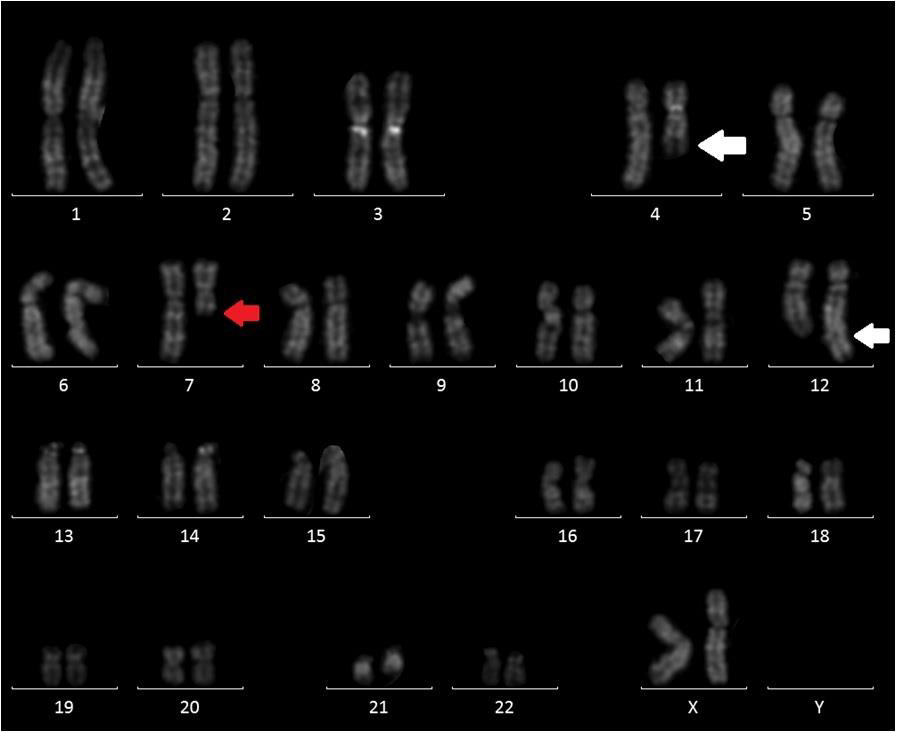
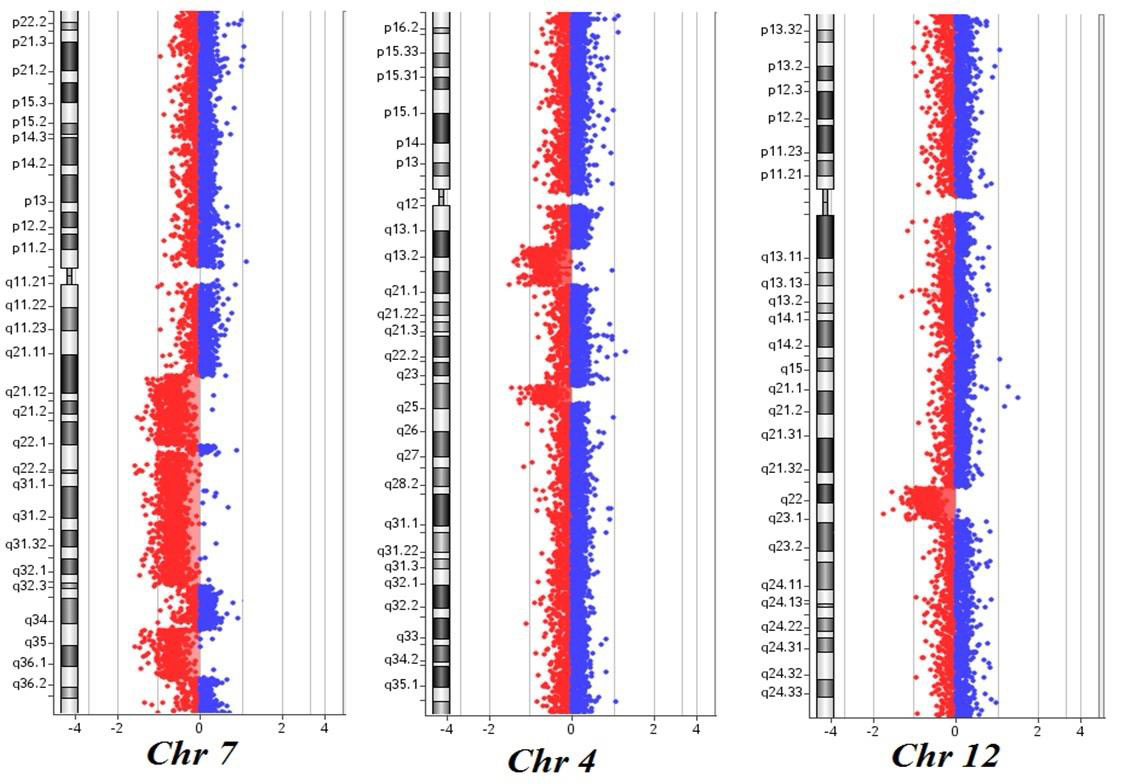
Here, we describe the case of a 76-year-old patient who was admitted to our hospital because of suspected CMML and for whom an array CGH was performed to better define the genomic imbalances at submicroscopic level and identify involved genes. In November 2018, a 76-year-old woman was referred to our hospital because of persistent monocytosis. A BM biopsy was then performed, showing increased age-adjusted cellularity and granulocytic proliferation associated with dyserithropoiesis and dysmegakaryopoiesis. A diagnosis of CMML-1, MD-subtype, was made according to the 2017 WHO classification. BM cytogenetic analysis revealed a karyotype characterized by the presence of two different cell lines, the largest one [18/20 metaphases] with an interstitial deletion of chromosome 7q at the bands q21-q36 and an apparently balanced translocation between chromosomes 4q24 and 12q15. Altogether, the karyotype was 46,XX,del(7)(q21q36),t(4;12) (q24;q15)[18]/46,XX[2] (Figure 1). A Cancer CGH+SNP array was then performed to define the real nature of the translocation. Array CGH analysis unveiled the t(4;12) unbalanced nature with three cryptic genomic imbalances: two deletions on chromosome 4 (one of 4.7Mb at band q24 spanning the bases 101944715-106679408, and one deletion of 10Mb at bands q13.1-q13.3, spanning the bases 64116915-74323464) and one deletion of 6Mb on chromosome 12 at bands q21.33-q23.1, spanning the bases 90077323-96215823 (Figure 2).
Figure 1: QFQ-banding abnormal karyotype of patient: white arrow showing the t(4;12) and red arrow the interstitial deletion of chromosome 7.
Figure 2: Cancer CGH+SNP array results of the patient: three noncontiguous deletions on chromosome 7q at bands q21.11-q22.1, q22.1-q32.2 and q34-q36.1; two deletions on chromosome 4 at bands q13.1-q13.3 and q24; one deletion on chromosome 12 at bands q21.33-q23.1. The breakpoints are according to the 37 build (March 2009) of the Human Genome Reference Consortium (GRch37/hg19).
Furthermore, the 7q deletion was composed of three noncontiguous deletions: a 15Mb loss at bands q21.11-q22.1, spanning the bases 82769585-98521920, a 30Mb loss at bands q22.1-q32.2, spanning the bases 100139536-130148949, and a 11Mb loss at bands q34-q36.1, spanning the bases 140529849- 151559567. Finally, the analysis did not detect any copy number neutral loss of heterozygosity. Based on these results, NGS analysis was then performed, showing the presence of TET2 c1870 (VAF 25.4%) and c3344 mutations (VAF 38.9%). These results are consistent with the presence of a normal cell line together with an abnormal one. As already reported in the literature, chromosome 7 aberrations are found in about 20% of CMML patients harboring cytogenetic abnormalities, classifying these cases as at high cytogenetic risk. On the long arm of chromosome 7 map several tumor suppressor genes and their loss of function via monoallelic deletion may play a role in CMML pathogenesis and progression. At present, tumor suppressor genes in 7q are believed to operate in a haplo insufficient manner, and new powerful technologies such as microarray comparative genomic hybridization allows to overcome this limit and new genes located in bands 7q22 and 7q34-36 have been discovered [6,7]. While chromosome 7q cytogenetic analysis could not detect the precise intervals and the genes involved in the deletion, with array CGH we identified five genes already known to have a potential role in tumorigenesis.
In details, EZH2 is a component of the polycomb repressive complex-2 and encodes for a methyltransferase, initiating epigenetic silencing of many genes involved in different cell pathways. CUX1 encodes for a homeobox transcription factor involving in tumorigenesis, with a possible role as a tumor suppressor gene. SAMD9 and SAMD9L compound heterozygous deletions with high frequency in adult and childhood myeloid leukemia. In contrast with previous reports, KMT2C/MLL3, despite being an epigenetic regulator acting as a gene silencer, is not involved in our deletion. In our patient, together with a del7q, we found an apparently balanced t(4;12) translocation, which was proved to be unbalanced by array CGH. The three deletions found on chromosome 4 involve many OMIM genes, with TET2 and NFKB1 playing an important role in disease progression. Somatic TET2 mutations occur in ~60% of CMML, even if they are not specific for the disease and can also be detected as a part of age-related clonal hematopoiesis. Moreover, they have not proven to negatively impact either on overall (OS) or leukemia-free survival [8,9]. On the contrary, in the absence of clonal ASXL1 involvement, TET2 mutations were shown to favorably impact on OS [10]. Interestingly, we found the coexistent loss of EZH2 due to the 11Mb deletion at bands q34-q36.1 of chromosome 7. Indeed, its deletion is known to contribute to myeloid tumorigenesis in association with TET2 variations. The 6Mb deletion of chromosome 12q involving 25 OMIM genes was not commonly described in association with hematological malignancies, so that its biological significance remains unclear. At the same time, we cannot exclude that some of the involved genes could play a minor role in disease onset or progression.
In conclusion, this case shows both common recurrent rearrangements and rare copy number alterations. Clarifying the role of these alterations could contribute to elucidate the mechanisms involved in CMML leukemogenic network, possibly contributing to define a more accurate prognosis. This case also underlines the importance of including different molecular cytogenetic tests in CMML diagnostic workup, so providing prognostic information and a strategy to develop personalized therapies, especially considering that NGS analysis is not always available.
Disclosure of Conflicts of Interest
All the authors declare they have no potential conflicts of interest.
Ethics Approval and Consent to Participate
This study was not conducted with research intervention. Thus,
ethics committee approval was not necessary.
The patient has given her written informed consent to publish
her case.
Funding
The only funds used were provided by the authors’ institutions.
References
- Patnaik MM, Tefferi A (2020) Chronic myelomonocytic leukemia: 2020 update on diagnosis, risk stratification and management. Am J Hematol 95(1): 97-115.
- Arber DA, Orazi A, Hasserjian R, Jurgen Thiele, Michael J Borowitz, et al. (2016) The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127(20): 2391-2405.
- Wassie EA, Itzykson R, Lasho TL, Olivier Kosmider, Christy M Finke, et al. (2014) Molecular and prognostic correlates of cytogenetic abnormalities in chronic myelomonocytic leukemia: a Mayo Clinic-French Consortium Study. Am J Hematol 89: 1111-1115.
- Elena C, Galli A, Such E, Meggendorfer M, Germing U, et al. (2016) Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood 128(10): 1408-1417.
- Ball M, List AF, Padron E (2016) When clinical heterogeneity exceeds genetic heterogeneity: thinking outside the genomic box in chronic myelomonocytic leukemia. Blood 128(20): 2381-2387.
- Jerez A, Sugimoto Y, Makishima H, Amit Verma, Anna M Jankowska, et al. (2012) Loss of heterozygosity in 7q myeloid disorders: clinical associations and genomic pathogenesis. Blood 119(25): 6109-6117.
- Inaba T, Honda H, Matsui H (2018) The enigma of monosomy 7. Blood 131(26): 2891-2898.
- Delhommeau F, Dupont S, Della Valle V, Chloé James, Severine Trannoy, et al. (2009) Mutation in TET2 in myeloid cancers. New Eng J Med 360: 2289-2301.
- Itzykson R, Kosmider O, Renneville A, Véronique Gelsi Boyer, Manja Meggendorfer, et al. (2013) Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol 31(19): 2428-2436.
- Patnaik MM, Tefferi A (2016) Cytogenetic and molecular abnormalities in chronic myelomonocytic leukemia. Blood Cancer J 6(2): e393.

 Case Report
Case Report