Research Article
Penetrance of Methylene Tetrahydrofolate Reductase
C677T Gene Polymorphism and Karyotypic Variations
Associated Increase Genetic Susceptibility in the Cases
of Congenital Heart Defects
Ajit K Saxena1*, Sanjeev Kumar2, Meenakshi Tiwari1 and Chandan Kumar Singh1
Author Affiliations
1Department of Pathology /Lab Medicine, All India Institute of Medical Sciences, India
2Department of Cardiothoracic and Vascular Surgery, All India Institute of Medical Sciences, India
Received:August 04, 2021 | Published:August 13, 2021
Corresponding author: Ajit Kumar Saxena, Professor and Head, Department of Pathology /Lab Medicine, All India Institute of
Medical Sciences, Patna - 801507 Bihar, India
DOI: 10.26717/BJSTR.2021.38.006103
Congenital heart disease (CHD), a multifaceted disorder occurs during embryogenesis
due to exposure of environmental mutagens (teratogens) exposed antenatally leading to
high-risk of infant mortality. Present study has been designed with the aims to evaluate
the frequency of chromosome variation and corelate to methylene tetrahydrofolate
reductase (MTHFR) C677T gene polymorphism as “risk factor” in clinically diagnosed
cases of CHD using lymphocytes cultures and ARMS PCR, respectively. FISH analysis
was carried for confirmation of chromosome-21. Interestingly, cytogenetics study
shows variation in the frequency of structural and numerical chromosome aberrations
with frequency in all the cases of CHD. Case-1, showing deletion of short arm of
chromosome-18 and Robertsonian translocation between G/G chromosome association
(24.00%), Case-2 showing numerical variation (trisomy-21), Case-3, includes dicentric,
chromatid break in chromosome-2, deletion of short arm in chromosome-5, reciprocal
translocation involving chromosome-6 and 10 and reporting first time appearance of ring
of Y-chromosome. Case-4 showing structural variations (16.00%) including dicentric,
chromatid breaks and trisomy of chromosome-21. The most common dominant
frequency was observed in karyotype trisomy-21(58.30%) in all the four cases of CHD as
an end point for genetic bio maker and showing significant differences (p < 0.001) using
X2- test between total number of chromosomes and trisomy-21 MTHFR (C677T) gene
polymorphism reveals (25.00%) of genetic heterozygosity of CT alleles and 75.00% cases
shows homozygosity of wild type (CC) alleles, suggesting the variations in the frequency
either in karyotypes or MTHFR C677T alleles are due to unconstitutional penetrance of
gene in the genome of CHD cases and increase genetic susceptibility to make the disease
more complex.
Keywords: Congenital Heart Disease;
Chromosomal Aberrations; FISH Analysis;
Trisomy-21; MTHFR Gene Polymorphism
Congenital heart defects (CHDs), the most severe congenital
anomalies associated with “Birth Defect” in infants and their
prevalence of 4 to 50 per 1000 live births and is associated with
high morbidity and mortality [1]. CHD shows multiple gene(s)
involvement during organogenesis including the environmental
factors [2-4]. Interestingly, approximately 30% of children of
CHDs shows chromosomal abnormality [1,3]. Among, the most
common cytogenetic abnormality is the origin of trisomy-21(Down
syndrome) in CHD cases [5]. Besides, chromosomal anomalies,
role of 5,10-methylenetetrahydrofolate reductase (MTHFR) gene
polymorphism associated DNA methylation has also been observed
earlier in CHD cases [6-11].
In human, MTHFR gene is thermolabile in nature and present
on chromosome 1p3 6.3. The polymorphic variation of C677T allele
(rs1801133) , the missense (point) mutation is responsible for the
reduction enzymatic activity (30% - 70%) followed by increase
“risk factor” in the variety of disease in heterozygous condition
other than CHDs, such as neural tube defects (NTD), mental
retardation, miscarriage and cancers [12-17]. Only few studies have
been demonstrated regarding variations of MTHFR allele in CHD
cases in Indian population [18,19]. Therefore, the present study has
been designed to evaluate the frequency of karyotypic variations in
individual case and try to find out their correlation with penetrance
of MTHFR C677T gene polymorphism as “risk factor” in CHDs cases
to confirm the hypothesis that congenital anomalies are polygenic
in nature.
Blood samples (1.0 ml) were obtained in sterile heparinised
vials from all the cases of CHD (n=4) and their respective controls
of the same age group after clinical diagnosis from the OPD of
Cardiothoracic and Vascular Surgery (CTVS) Department and
referred to Human Molecular Cytogenetic Laboratory of the
Department of Pathology / Lab Medicine to evaluate the causative
genetic risk factors, after clearance form Institute Ethical Committee
(IEC), All India Institute of Medical Sciences, Patna.
Lymphocytes cultures were set up in the media (RPMI 1640)
supplemented with foetal calf serum (5%), Phytohaemagglutinin-M
and antibiotic (penicillin & streptomycin) for 72hrs in CO2 incubator
at 37°C under sterile conditions. Harvesting was performed
after adding colchicine (10 μl) in the cultures to arrest mitosis.
Prewarmed KCL (0.056%) solution was used as hypotonic, and cells
were fixed in acetic acid and methanol (1:3) as detailed procedure
is illustrated earlier by Saxena et al in 2020. Giemsa stain was used
for staining and GTG banding was performed after using (0.001%)
trypsin in PBS buffer (pH 7.4) and karyotyping was performed
according to the ISCN (2016) [20] using applied spectral imaging
software (Genesis, USA).
FISH analysis was carried out for the study of D21S65 specific
region for chromosome-21 in both interphase and metaphases
plates after using probe LSI 21 spectrum orange having region
21q22.13-q22.2 of 220 kb, obtained from Abbott-Vysis, Inc. (USA).
Chromosome - 21 was identified by green signal and nuclear DNA
was labelled with DAPI as counter stain (Blue).
MTHFR C677T gene polymorphism was carried out by RT
RT-PCR (Bio Red, USA) using Syber green as fluorescence dye,
procedure published earlier by our research group21. Genomic
DNA were isolated using Promega DNA isolation Kit (USA)
using manufacturer protocol and quantified using Nanodrop
spectrophotometer (Thermo scientific, USA). The samples were
stored at -20o C till the PCR reaction was performed. MTHFR
677CT primers were designed for tetra plex RT PCR assay (http://
cedar.genetics.soton.ac.uk/public_html/primer1.html) and BLAST
program at http://www.ncbi.nlm.nih.gov/blast to determine the
specificity of the primers. To increase the specificity of the reaction
a mismatch at the 2 positions of the 3’ end both the allele-specific
primers were selected and confirmed by software. The selection of
the primers was based on the amplicons ‘Tm’ values and following
primers used in present study:
MTHFR-T,5’–GCACTTGAAGGAGAAGGTGTCTGCGGGCGT-3’.
MTHFR-C-polyG,5’-GGCGGGCGGCCGGGAAAAGCTGCGTGATGAT
GAAATAGG-3’.
MTHFR-cf,5’-TGTCATCCCTATTGGCAGGTTACCCCAAA-3’.
MTHFR-cr,5’- CCATGTCGGTGCATGCCTTCACAAAG-3’.
To obtain amplicons with distinct melting points, ‘Tm’ values
were calculated using known software (http://eu.idtdna.com/
analyzer/Applications/OligoAnalyzer/). We have plan to select
these primer ARMS PCR based on “Tm” analysis, part of plan of
our interest to detect SNP of mutant of MTHFR allele(s), same
PCR product was further confirmed by agarose gel (2.0%)
electrophoresis and bands were visualized and characterized after
ethidium bromide staining on Gel Doc system.
Simple procedure includes a total volume of 20 μl containing 10
μl of SYBR Green PCR Master Mix, 1 μl of each primer per reaction,
40 ng of genomic DNA, and distilled water was taken to perform
RT-PCR. The PCR protocol on the light cycler included an initial
denaturation step (95 0C for 7 min) was followed by amplification
and quantification steps repeated for 30–40 cycles (950C for 10 s,
600C for 10 s, 720C for 20 s), with a single fluorescence measurement
at the end of the elongation step at 720C, reaction was terminated
by cooling to 400C. Melting curves were constructed by lowering the
temperature to 650C and later increasing the temperature by 0.20C/s
to 980C to measuring the change in fluorescence consistently. Tm
values were assigned to develop plot generated by the RT-PCR of
the negative derivation of fluorescence versus temperature (dF/
dT) of the melting curve for amplification products measured at
530 nm.
Chi square (X2) test was used to find to calculate p value and
level of significance (p=value) between normal and abnormal cells
during analysis of karyotypes in individual case of CHDs.
Cytogenetics analysis in CHD cases were performed for
developing karyotypes individual to find end point according to
ISCN 2016. The cytogenetic findings include a spectrum of GTG
karyotypes showing large number of structural and numerical
variations in individual cases of CHDs with brief history. The detail
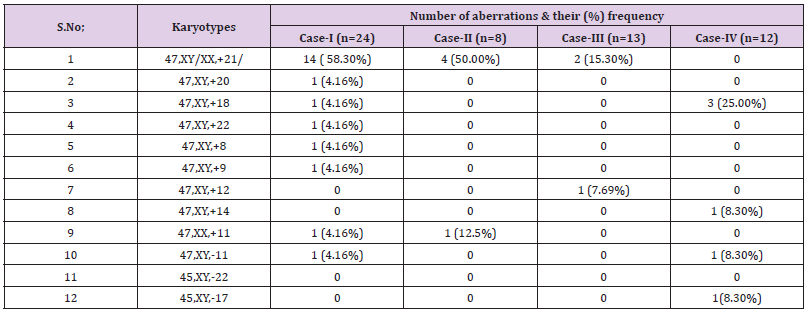
findings of numerical as well as structural variations (% frequency)
in individual cases were documented in Table 1.
Case-I
Three months old male child, clinically diagnosed CHD based
on findings of ECHO cardiogram with AV+VA concordance and RA/
RV mildly enlarged. The cytogenetic study showing dominantly of
47 XY+21, confirmed down syndrome with the frequency of 58.3%.
Other trisomy identified were 47 XY +20 karyotype (4.16%), 47
XY+18 (4.16%), 47 XY +22 (4.16%), 47 XY +9 (4.16%), 47 XY+8
(4.16%), 47XY+11 4.16%); 45 XY -11 (4.16%).
Case-II
The one and half month-old, female child presented with CHD
with clinical features including abnormal echocardiogram showing
VSD (having thickness of 0.5 cm) with clinical features of Down’s
syndrome The frequency of numerical variations having trisomy -
21 (47 XX+21) were 50% and 47 XX+11 was 12.5%.
Case-III
This case includes male, age two year and one month have AV
malformations with multiple venous malformations in chest and
abdominal wall with significant fatty components and diffusely
infiltrated muscles (angiography). Karyotypes shows 15% of
cells with trisomy - 21 (47 XY+21) while trisomy -12 (47 XY+12)
is 7.7%. In this case the structural chromosomal abnormities are
dominant and shows more than 30%, such as chromatid breaks of
chromosome - 2 (2p) and chromosome 10 (10q) and formation of
ring of Y chromosome. Besides these structural abnormalities one
metaphase showed centromere breakage (> 60%) in chromosomes.
Case-IV
(P-593), Clinically diagnose on the bases of echocardiogram
showing Situs Solitus, AV/VA concordance, Chamber size normal,
valves normal. Cytogenetics findings includes 47 XY +18 (25%);
47XY+14 (8.3%), 45 XY-11 (8.3 %) and 45XY -17 (8.3%). Structural
aberrations showing more than 8% chromatid breaks involving
chromosome 1(1p23).
The variations in the frequency of chromosomes due to different
clinical conditions and severity of disease. Interestingly, numerical
variations and their frequency are predominantly dominated in
all the cases of CHD including in the karyotypes of trisomy-21, 18,
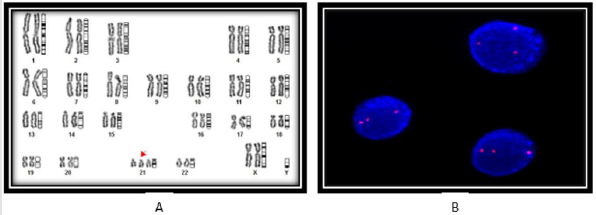
and monosomy of chromosome -11, a representative karyotype
showing trisomy-21 is shown (Figure 1A). Simultaneously,
structural chromosomal aberrations including chromatid breaks in
chromosome -1(-1q) and 2 (-2p), besides this formation of Y-ring
chromosome and centromeric breakage were the striking feature
in two cases of CHD with frequency > 8% to 30%, respectively.
The numerical chromosomal variations were observed in all the
four cases with maximum frequency (58.30%) were observed in
karyotypes 47, XY + trisomy 21. The extra copy of chromosome-21
was further confirmed by FISH analysis using specific probe LSI 21
spectrum orange having region 21q22.13-q22.2 of 220 kb as shown
in Figure 1B. Interestingly, only one case showing lack of trisomy -
21, but extra copy of chromosome -18, 14 (trisomy) was observe
with monosomy of chromosome -11, 17, respectively.
Single nucleotide polymorphism (SNP) of MTHFR gene regulate
folate metabolism is important event during embryogenesis. Highly
sensitive, allele refractive mutation specific (ARMS) PCR based
technique is used for SNP analysis to determine “risk factor” of
MTHFR C677T genotyping - wild-type (CC) allele and mutant (TT)
allele in homozygous condition and heterozygous condition (CT).
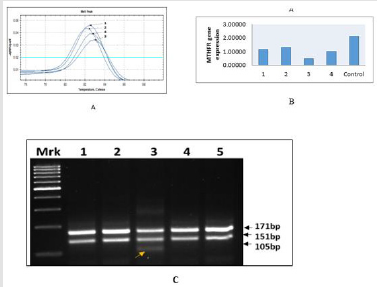
In all the cases of CHDs, the Tm value showing shift from 82.50C
to 83.00C, suggesting increase heterozygocity (CT) in 25% of cases
due to allele C change into T (C→ T) as mentioned in Figure 2A,
suggesting increase “risk” of the disease, while rest of the three
cases (75%) showing only wild type of genotype CC (Figure 2A-
2C). The findings RT-PCR were further confirmed the appearance of
additional band of 105 kb belong to mutant “T” allele (Case - 3) on
agarose gel electrophoresis as documented in Figure 2C.
The etiopathology of CHDs are complex due to the interactions
of genetic and environmental factors [4,22]. It has been identified
that ~30% of CHD cases are syndromic involving chromosomal
aberrations, while non-syndromic are monogenic and shows variety
of genes. Now it is evident form preview of the fact that genetic
factors become essential to understand the etiopathology in CHD
cases and risk assessment before initiation of clinical management
and counselling. However, there is lack of the genetic knowledge
of CHDs in Indian population, only few studies have been achieved
to study an association of genetic factors in CHDs. Therefore,
present study was performed to accumulate the data from different
approaches like karyotypic pattern and MTHFR (C677T) gene
polymorphism to determine “risk factors” in CHD patients.
Epidemiological studies revel that frequency (%) and types of
chromosomal abnormalities varies between USA or Europe, but
there is a lack of data in Indian population. Hence, the present study
on CHDs becomes relevant to assess the risk using MTHFR C677T
gene polymorphism analysis and try to correlate with karyotypic
variations with agreement by the authors present study is small,
but quite interesting. Previous studies identified chromosomal
abnormalities with variation in the frequency from 3-18% in
CHDs due to different variables such as clinical characteristics, age
and ethnicity [22, 23]. In the present study, numerical variations
(trisomy-21) show very high frequency (58.30%) and dominant
over all kind of structural and numerical variations, the detail data
of cytogenetics is documented in tabular form (table-I). Present
findings are the agreement of earlier studies that trisomy-21 and
18 are associated with the cases of CHD [23,24]. Previous stud
also reported trisomy-13, however, such chromosomal aberrations
failed to observe in the present study may be either due to small
sample size or different environmental factors [24].
In the present study, frequency of structural chromosomes was
varying from 8-30% alone over total cytogenetics alterations and
more interesting than numerical end point due to the appearance of
“Y-ring chromosome and centromeric breakage reporting first time.
Author failed to interpret the present findings that there might
be involvement of conserve DNA sequences exist at centromeric
regions and role of sex-chromosome during pathogenesis of
CHD cases. Earlier study shows the deletion of the short arm of
chromosome - 6 and 17, while present study also documented
chromatid breaks - unstable regions of chromosomes [22-25].
Interestingly, present study shows “novel” structural changes has
not been reported earlier with high impact on genomic instability
of CHDs genome.
The conversion of folate 5,10-methylenetetrahydrofolate to
5-methyltetrahydrofolate in multistep processes that converts
homocysteine to S-adenosyl methionine involving methyl donor
for the maintenance of several biological processes. Methionine
is incorporated into protein synthesis, growth and metabolism
processes. The S-adenosyl methionine serves as a role for clearance
of harmful substances, metabolites and waste products more
efficiently. Earlier study shows that deficiency in MTHFR activity
leads to increase homocysteine and lowering of folate levels in
circulation. Homocysteine, is a Sulphur containing amino acid, acts
as biomarker of “risk factor” of cardiovascular disease by promoting
oxidation of lipids and lipoproteins [26]. Growing number of
evidence also suggest that mutations in folate metabolism may also
be involved in genetic susceptibility to CHD. In 2001, Junker and
colleagues showed that significant association of MTHFR 677TT
genotype in the development of CHDs during early pregnancy [8-11,
27]. There is still needed to explore new variants of MTHFR and their
association to the “risk factor” during organogenesis in developing
countries like India needs to be explored. In the present study, we
report for the first time the role of MTHFR C677T polymorphism
and codominance of chromosome variations in CHDs cases in the
Eastern region of India. Previously, few studies have been reported
mutant “T’’ allele of MTHFR in CHDs [18,19]. Our results indicate
that the frequency of the putative risk allele C677T was 25%,
which is similar to meta-analysis study performed by Xuan et al
where they identified frequency of 28.99% in Caucasian children
and 31.76% in the Caucasian maternal population, although higher
frequency (42.28%) has been reported in Asian population [28].
Importantly, studies by Hobbs and co-workers identified Hcy,
S-adenosylhomocysteine, and methionine as the most important
biomarkers predictive in mothers whose pregnancies were affected
by congenital heart defects (224 case subjects) or unaffected
by any birth defect (90 control subjects)7. The variations in
allele frequency may be either due to different clinical feature
or penetrance of mutant allele to the CHDs genome to increase
severity of the disease, but simultaneously the epigenetic factors
should not be ignored.
From the present study we are reporting first time the
penetrance of MTHFR C677T gene polymorphism as “risk factor”
with karyotypic variations in the cases of CHDs. Although, this study
is based on small group, but findings are promising with new findings
in chromosome aberrations are documented as independent risk
factor during embryogenesis, suggesting to increase large samples
size for validity of the findings in other groups. However, other
than genetic factor influences by consanguinity and environmental
factors (socioeconomic) together cause genomic instability
resulting abnormal development of foetus such as CHDs.
AKS thankfully acknowledges to the Director, AIIMS Patna also
give special thanks to the patients who participates in the study.
All the authors have agreed with the content of the manuscript.
There is no conflict of interest between the authors.
AKS and MT are involved for genetics analysis, SK is responsible
for clinical diagnosis and management of the patients, while CK
help during preparation of the manuscript.
- Mary Ella Pierpont, Craig T Basson, D Woodrow Benson Jr, Bruce D Gelb, Therese M Giglia, et al. (2007) Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115(23): 3015-3038.
- Richards AA, Garg V (2010) Genetics of congenital heart disease. Curr Cardiol Rev 6(2): 91-97.
- Chaix MA, Andelfinger G, Khairy P (2016) Genetic testing in congenital heart disease: A clinical approach. World J Cardiol 8(2): 180-191.
- Tian G, Gao H, Hu S, Shu Q (2018) Research progress on genetic and epigenetic mechanisms in congenital heart disease. Zhejiang Da Xue Xue Bao Yi Xue Ban 47(3): 227-238.
- Benhaourech S, Drighil A, Hammiri AE (2016) congenital heart disease and Down syndrome: various aspects of a confirmed association. Cardiovasc J Afr 27(5): 287-290.
- Zhu WL, Li Y, Yan L, Dao J, Li S (2006) Maternal and offspring MTHFR gene C677T polymorphism as predictors of congenital atrial septal defect and patent ductus arteriosus. Mol Hum Reprod 12(1): 51-54.
- Charlotte A Hobbs, Mario A Cleves, Stepan Melnyk, Weizhi Zhao, S Jill James (2005) Congenital heart defects and abnormal maternal biomarkers of methionine and homocysteine metabolism. Am J Clin Nutr 81(1): 147-153.
- Chrysovalanto Mamasoula, R Reid Prentice, Tomasz Pierscionek, Faith Pangilinan, James L Mills, et al. (2013) Association between C677T polymorphism of methylene tetrahydrofolate reductase and congenital heart disease: meta-analysis of 7697 cases and 13,125 controls. Circ Cardiovasc Genet 6(4): 347-353.
- Wang W, Wang Y, Gong F, Zhu W, Fu S (2013) MTHFR C677T polymorphism and risk of congenital heart defects: evidence from 29 case-control and TDT studies. PLoS One 8(3): e58041.
- Martí-Carvajal AJ, Solà I, Lathyris D, Dayer M (2017) Homocysteine-lowering interventions for preventing cardiovascular events. Cochrane Database Syst Rev 8(8): CD006612.
- Zhang R,Huo C, Wang X,Dang B, Mu Y, et al. (2018) Two Common MTHFR Gene Polymorphisms (C677T and A1298C) and Fetal Congenital Heart Disease Risk: An Updated Meta-Analysis with Trial Sequential Analysis. Cell Physiol Biochem 45(6): 2483-2496.
- Saxena Ajit K (2011) MTHFR C677T gene mutation associated with severe risk for mental retardation in children. Int J of Genetics 3(2): 47-49.
- Gupta G, Gupta J, Pandey S, Pandey LK, Saxena Ajit K (2011) Role of Cell free fetal DNA in maternal blood - a prospective role of CβS & MTHFR gene as antenatal genetic marker in pre-eclampsia patients. Int. J. of Genet 3(2): 62-65.
- Saxena Ajit K, Pandey S, Pandey LK (2012) Evaluation of Methylenetetrahydrofolate reductase C677T gene polymorphism associated risk factor in the patients of recurrent pregnancy loss. J. Med. Genet. Genomics 4(2): 25-28.
- Singh A, Pandey S, Pandey LK, Saxena Ajit K (2015) In Human allele specific variation of MTHFR C677T and A1298C associate risk factor for the Development of Ovarian Cancer. J. Exp. Therap. Oncol 11(I): 67-70.
- Saxena Ajit K, Tiwari M, Kumar V, Singh CK (2020) MTHFR New Gene Variants Increase Risk Factor in Wilms’ tumor and Prediction of 3D Structure Modulates Functional Activity During Drug-Protein Interaction. J of Integrative Oncol 10: 1.
- Saxena Ajit K, Agarwal M, Kumar A, Singh CK (2021) Genetic heterogenecity of MTHFR C677T allele modulate hormonal dysfunction associated risk factors in the cases of male infertility. Int J Dev Res 12: 1-6.
- Mukherjee M, Joshi S, Bagadi S, Dalvi M, Rao A, et al. (2002) A low prevalence of the C677T mutation in the methylenetetrahydrofolate reductase gene in Asian Indians. Clin Genet 61(2): 155-159.
- Vasisht S, Gulati R, Narang R, N Srivastava,Lalit Mohan Srivastava, et al. (2002) Polymorphism (C677T) in the 5,10-methylenetetrahydrofolate reductase (MTHFR) gene: A preliminary study on north Indian men. Indian J Clin Biochem 17(1): 99-107.
- Shaffer, Lisa G, Schmid M, McGowan-Jordan (2016) Iscn: An International System for Human Cytogenomic Nomenclature (2016). Basel: Karger.
- Saxena Ajit K, Gupta RK, Kumar M (2016) ARMS-PCR based SNP analysis of MTHFR C677T allele using Syber green in pancreatic tumor. British Journal of Medicine and Medical Research 11(12): 1-6.
- Robert J Hartman, Sonja A Rasmussen, Lorenzo D Botto, Tiffany Riehle-Colarusso, Christa L Martin, et al. (2011) The contribution of chromosomal abnormalities to congenital heart defects: a population-based study. Pediatr Cardiol 32(8): 1147-1157.
- Patrícia Trevisan,1 Tatiana Diehl Zen, Rafael Fabiano Machado Rosa,Juliane Nascimento da Silva,Dayane Bohn Koshiyama, et al. (2013) Chromosomal abnormalities in patients with congenital heart disease. Arq Bras Cardiol 101(6): 495-501.
- C Ferencz, C A Neill, J A Boughman, J D Rubin, J I Brenner, L W Perry (1989) Congenital cardiovascular malformations associated with chromosome abnormalities: an epidemiologic study. J Pediatr 114(1): 79-86.
- Paskulin GA, Zen PR, Rosa RF, Manique RC, Cotter PD (2007) Report of a child with a complete de novo 17p duplication localized to the terminal region of the long arm of chromosome 17. Am J Med Genet A 143A (12): 1366-1370.
- P Frosst, H J Blom, R Milos, P Goyette, C A Sheppard, et al. (1995) A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet 10(1): 111-113.
- R Junker, S Kotthoff, H Vielhaber, S Halimeh, A Kosch, et al. (2001) Infant methylenetetrahydrofolate reductase 677TT genotype is a risk factor for congenital heart disease. Cardiovasc Res 51(2): 251-254.
- Chao Xuan, Hui Li, Jin-Xia Zhao, Hong-Wei Wang, Yi Wang, et al. (2014) Association between MTHFR polymorphisms and congenital heart disease: a meta-analysis based on 9,329 cases and 15,076 controls. Sci Rep 4: 7311.

 Research Article
Research Article