Abstract
Purpose: The Renin Angiotensin Aldosteron System (RAAS) is one of the major systems involved in the pathophysiology of heart failure. In this study we investigate the association between the renin angiotensin aldosteron system genes polymorphisms and acute heart failure (AHF) and we evaluate the role of serum ACE activity.
Methods: We included 300 patients over 20 years old admitted to the Emergency Department for acute dyspnea. According to the clinical findings and results of the B-type natriuretic peptide (BNP level), patients were divided into two groups: heart failure (HF) and non- heart failure (non-HF) groups. Genotyping was done by PCR-RFLP. The level of serum ACE activity was determined by spectrophotometric method using hippuryl histidyl leucine. We found the diagnosis of AHF in 50% of the population.
Results: No association between the renin polymorphism and AHF, but we found an association between ACE ID and CYP11B2 polymorphisms and heart failure. Level of serum ACE activity is significantly higher in the group of AHF.
Conclusion: We can conclude that blood ACE activity, ACE ID and CYP11B2 can be a marker of acute heart failure.
Keywords: Acute Heart Failure; Renin; ACE ID; Serum ACE Activity; Aldosteron; Polymorphisms
Abbreviations: HF: Heart Failure; RAS: Renin–Angiotensin System; ACE: Angiotensin- Converting Enzyme; EDTA: Ethylene Diamine Tetra Acetic Acid; PCR: Polymerase Chain Reaction
Introduction
Heart Failure (HF) is an ever increasing problem worldwide and is a major health problem associated with very high morbidity and mortality [1]. It can be defined as a complex clinical syndrome that results from any structural or functional disorder of the heart to achieve the ability of the ventricles to fill or eject blood [2]. The main events of the HF is dyspnea and fatigue, that limit exercise tolerance and induce water retention that may lead to pulmonary congestion and peripheral edema [3]; these two anomalies hinder the functional capacity and quality of life of those affected [4]. Acute heart failure may result from the development of a variety of arrhythmias; acute coronary syndrome, a rapid increase in the work of a failing heart by conditions such as infection, anemia, and pulmonary embolism [5], discontinuation of treatment of chronic HF and progression of the underlying disease [6]. High blood pressure can also bring on heart failure by causing left ventricular hypertrophy [7]. The Renin–Angiotensin System (RAS) is a central regulator of cardiovascular and renal functions and plays an important role in the pathophysiology of heart failure [8,9]. Renin, secreted by the kidneys, will convert angiotensinogen to angiotensin I, which then will become angiotensin II. Then aldosterone will be released from the adrenal cortex, leading to sodium and water retention and an increase in circulating blood volume. In the persistence of this mechanism without control, it can aggravate heart failure as the heart struggles to pump against the increased volume. Genetic deletion of β-arrestin-1 (achieved by a global knockout mouse model) markedly improved cardiac function, adverse remodeling, aldosterone levels, and cardiac β Adrenergic Receptors function during HF progression. ((Hypertension. 2014;63:215-6).
The deleterious effect of this kinase on β adrenergic receptor signaling has been also explored in mice, showing that GRK2 gene deletion could prevent the development of post-MI HF and restore heart function (Am J Cardiol. 2011 Apr 15;107:1125-30). Given the important homeostatic actions of the RAAS on the cardiovascular system, it is not surprising that the synthesis and secretion of (pro- ) renin are tightly controlled [10]. To date, few published studies have examined the association between the polymorphism of this system and HF. Renin is the key enzyme at the top of the Angiotensin RAS, it cleaves the N terminus of circulating angiotensinogen to angiotensin I, which is then transformed into Angiotensin II by soluble or endothelial cell–associated Angiotensin-Converting Enzyme (ACE) [11]. The ACE ID polymorphism, on intron 16, results from the insertion (I) or deletion (D) of a 287-base pair Alu sequence [12] and the D allele has been linked to an increased plasma ACE activity [13], and a higher risk for heart failure [14]. The association between the polymorphism of the renin gene C-4063 T and HF has not been investigated previously. On the other hand, the aldosterone synthase gene, CYP11B2, encodes for a cytochrome P450 enzyme, involved in the terminal steps of aldosterone synthesis in the zona glomerulosa cells of human adrenal glands and its expression is regulated by angiotensin II and potassium [15].
The candidacy for this gene is based on its pathogenic role in the syndrome of glucocorticoid-remediable aldosteronism [16]. Aldosterone excess is a well-documented cause of hypertension and there is convincing evidence that mineralocorticoids have adverse effects in heart failure [5].The key enzyme in aldosterone synthesis is aldosterone synthase. The corresponding gene CYP11B2 is located on chromosome 8. A cytosine/thymidine (C/T) substitution in the 5’ promoter region at location _344 of the CYP11B2 gene has been identified. The functionality of this variant is unclear [1]. Several common polymorphisms have been described in the CYP11B2 [17]. The C-344 T polymorphism, which is located at a putative binding site for the steroidogenic transcription factor (SF- 1), has been associated with hypertension [18]; this locus may be important in blood pressure and cardiovascular regulation [17]. In the present study, we investigated the association, for the first time, between renin, ACE ID and aldosterone polymorphisms in North African patients admitted to the emergency department for dyspnea and AHF and we evaluated the role of serum ACE activity.
Methodology
Subjects and study design: This is a prospective descriptive study in the emergency department of Monastir. We included all patients aged over 20 years consulting in the emergency for acute dyspnea. We excluded all patients with cardio-respiratory arrest, coma and shock. At the time of study entry, family history, physical examination findings, medical therapy, and results of prior cardiac testing were collected. Patients were assigned to two groups: the HF Group: if the level of pro NT pro-BNP is greater than or equal to 1600pg / ml. The non HF group: if the level of pro NT pro-BNP is less than 400 pg/ml. For intermediate values: patients were classified in the HF group or non HF after consulting two experts (one emergency physician and one cardiologist) who established the diagnosis of HF on the basis of clinical data, NT pro-BNP levels and echocardiography findings. Venous blood samples were collected from every patient (4 ml EDTA tube for DNA extraction and 4ml in heparinized tube for the biochemical tests).
Biochemical Measurements
Serum total cholesterol, HDL-C and triglycerides were measured using enzymatic methods on Beckman autoanalyzer (Randox, Antrim, UK). LDL-C cholesterol was calculated using Friedewald’s formula. NT-proBNP was measured by the electrochemiluminescence assay (Elecsys 2010, Roche Diagnostics, Germany). Biochemical serum parameters (creatinin, uric acid, glycemia) were determined on Cobas 6000™ (Roche Diagnostics).
ACE I Activity
The spectrophotometric method using Hippuryl Histidyl Leucine (HHL), describes an enzymatic assay and detection by ultraviolet ACE I blood. This is the original method of Cushman and Cheung, 1971. Angiotensin converting enzyme is a glycoprotein peptiylpeptide hydrolase which section the histidyl leucine dipeptide angiotensin I in a relatively inactive decapeptide. The reaction is carried out at 37 °C and pH 8.3. The substrate (HHL) is transformed by the action of ACE histidyl leucine + hippuric acid.
ACE
Hippuryl-histidyl-leucine hippurate+ histidyl-leucine
After extraction of hippuric acid with ethyl acetate hippurate assayed by reading the optical density at a wavelength of 228 nm.
Genotyping Methods
Five milliliters of venous blood was collected with Ethylene Diamine Tetra Acetic Acid (EDTA) as anticoagulant. The genomic DNA was extracted from the peripheral leucocytes using the standard phenol-chloroform extraction method.
ACE ID Polymorphism
ACE ID polymorphism was determined by RFLP Polymerase Chain Reaction (PCR). Reactions were performed with 12.5 pmoles of each primer: sense oligo 5´ CTG GAG ACC ACT CCC ATC CTT TCT 3´and anti-sense oligo: 5´GAT GTG GCC ATC ACA TTC GTC AGA T 3´ in a final reaction volume of 25 μL, containing 3mM MgCl2, 400 μM of each dNTP , 50 mM KCl, 10 mM Tris-HCl (ph 8.5) and 1 unit of Taq DNA Polymerase. The DNA was amplified for 30 cycles with denaturation at 94 0C for 1 min, annealing at 65 0C for 1 min, and extension at 72 °C for 1 min using. PCR products were electrophoresed in 2% agarose gel and analyzed by ethidium bromide stain under ultraviolet light. Genotype interpretation was based on the length of the PCR products: 190 and 490 base pairs for the deletion (D) and insertion (I) alleles, respectively. To avoid mistyping of ID subjects as DD due to insufficient amplification of I allele, all samples genotyped as DD by the standard amplification procedure were then subjected to a second independent PCR amplification with a primer pair that recognizes an insertionspecific sequence (5’-TGG GAC CAC AGC GCC CGC CAC TAC-3’; 5’-TCG CCA GCC CTC CCA TGC CCA TAA-3’), with identical PCR conditions except for an annealing temperature of 670C.
Renin C-4063T Polymorphism
Renin C-4063T polymorphism was using the following primers: (sense) 5′- AAA CTA GAA TGG GCT ACC AGA-3′, and (antisense): 5′- GCT GTG ACT TGT CTC TTC CTG A–3′; with the C (163 and 68 bp) and T (235 bp) alleles visualised on 2.5% agarose gel. Briefly, for a 25 μL PCR final volume, the reaction contained 200 ng genomic DNA, 250 μmol/l of each of dNTPs (dATP, dCTP, dGTP and dTTP), 250 ng of each primer, 1.5 mmol/l magnesium chloride and 1 U Taq DNA polymerase. PCR amplification consisted of an initial 1 min denaturation at 94 °C, followed by 40 cycles of denaturation at 94 °C for 40 s, annealing at 52 °C for 40 s, and extension at 72 °C for 40 s. The terminal extension was performed at 72 °C for 5 min. The PCR product was digested with the Taq I restriction enzyme (Promega). Digested products were separated by agarose gel electrophoresis and visualized directly under UV light after staining with ethidium bromide. Undigested 235 bp fragment indicated the presence of the T allele and, appearance of two bands at 163 and 168 bp represented the C allele.
C-344T CYP11B2 Polymorphism
PCR conditions were initial denaturation at 94 °C for 2 min; then, 30 cycles at 94 °C for 30 s, at 67 °C (annealing) for 30 s, at 72 °C (extension) for 30 s; final extension at 72 °C for 5 min. Subjects were genotyped for the -344 promoter polymorphism using primers CAGGAGGAGACCCCATGTGAC (sense) and CCTCCACCCTGTTCA GCCC (antisense). Restriction Fragment Length Polymorphism Analysis (RFLP) was performed by adding 10 U of restriction endonuclease Hae III (Gibco BRL) in the appropriate buffer to 5 _l from each reaction (a 537-bp product) and by incubating at 37 °C for 2 h. The samples digested then underwent electrophoresis on 2.0% agarose gel with a Gel Electrophoresis, ethidium bromide stained, and analysed under UV-light. Since the -344T allele lacks an Hae III site (GGCC) present in the -344C allele, the -344T alleles are detected as fragments of 273 bp and -344C alleles as fragments of 202 bp.
Statical Analyses
The SPSS version 18.0 program was used for the statistical analysis, and the significance level was set at 5%. The Pearson chi-square test was used to determine the association between categorical variables. The odds ratio (OR) and their 95% Confidence Interval (CI) were also calculated. Differences between frequencies were assessed using the chi-square test, with a p-value of 0.05 or less was considered as significant.
Results
Study subjects
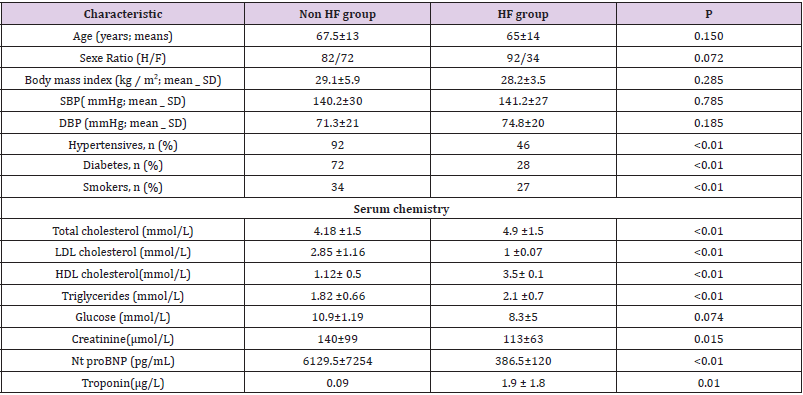
We included 300 patients, a HF group (n=150; 50%); and a Non HF group (n=150; 50%). The characteristics of study participants are shown in Table 1, with a mean age of 67.5 ±13 years and a mean left ventricular ejection fraction of 59% for the Non HF group and 46% for the HF group. All patients were NYHA (New York Heart Association) class III or IV at the time of admission. Clinical exams findings were similar between the two groups. Compared with the Non-HF patients, HF patients showed a significantly higher prevalence of conventional risk factors for heart failure, including high blood pressure, diabetes and smoking. The HF group also showed significantly higher levels of NT-pro BNP, lipids and troponin.
Genotype Analysis
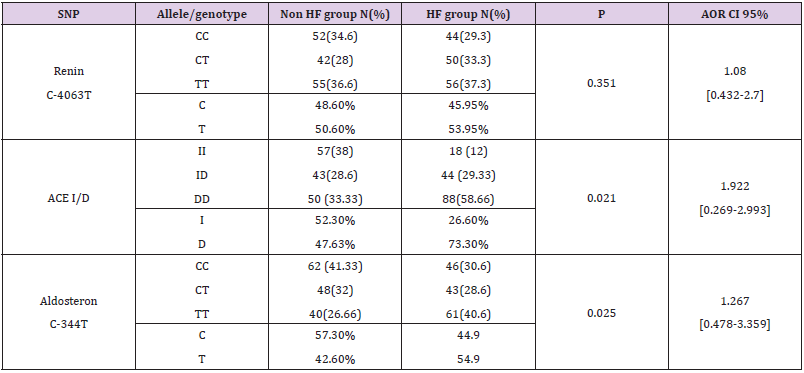
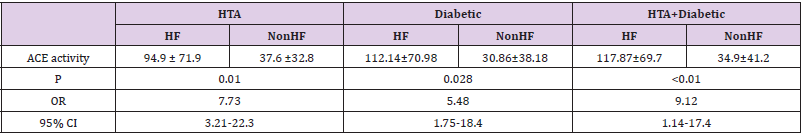
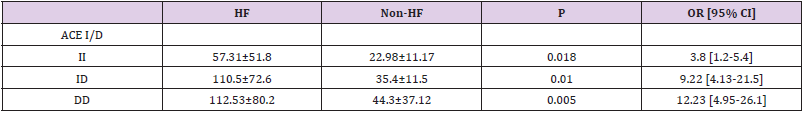
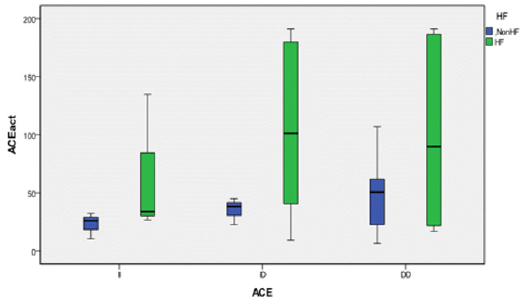
We genotyped all patients included in this study for renin, ACE and Aldosteron polymorphisms. Genotypes at all genes were in the Hardy–Weinberg equilibrium. Association was tested for each polymorphism separately. Results of analyses for single polymorphisms are shown in Table 2. The allele and genotype distribution of renin C-4063 T SNP is comparable between the Non HF and the HF groups (p=0.351). Whereas we found an association between HF and ACE ID and Aldosteron polymorphisms with p=0.021 and p=0.025, respectively. ACE ID polymorphism and serum ACE Activity. Compared with the different Risk Factor, serum ACE activity is higher in the group of patients who has heart failure associated with hypertension and diabetic with a higher risk (OR=9.12, CI 95% [1.14-17.4]) (Table 3). The I allele had a lower level of serum ACE activity compared with DD genotype. The frequency of DD genotype and D allele was significantly higher in HF group compared to the Non HF group and was associated with increased risk of HF (Table 4, Figure1). The DD genotype have significantly higher serum ACE activity compared to ID and II (p<0.01). As depicted in figure 1, ROC curve analysis displayed an area under the curve for the diagnosis of AHF. The AUC of the ROC curve for ACE activity indicated 0.709.
Discussion
Angiotensin-Converting Enzyme ID Polymorphism
The ACE polymorphism has a potential role as a modifier gene in heart failure; in the present study, we found an association between ACE ID polymorphisms and a higher risk of HF (p=0.021, OR=1.922, CI 95% [0.269-2.993]), furthermore, the DD genotype is associated with the risk of HF in our study’s population. Like our finders, many studies have found that the DD genotype can be associated with the risk of HF in Swedish patients [19], and Caucasian’s patients [20] also it can reduce the cardiac function and increased cavity size in South African patients with cardiomyopathy [21]. Another study on Chinese patients, despite they confessed that the study’s population was not in Hardy-Weinberg equilibrium, approved the presence of this association [22]. Most studies did not find an association between the ACE polymorphism and heart failure. Bleumink and colleague did not find an association to ischemic or idiopathic dilated cardiomyopathy with the ACE ID polymorphism [1]. Yamada et al. also did not detect an association between ACE genotypes and hypertrophic cardiomyopathy in Japanese, neither in Caucasian study published by Marian [23] and a second one where Bilsel confirmed the association between the D allele and dilated cardiomyopathy in Caucasian population [24]. These results also were confirmed on a Chinese population [25] and black South African subjects [26,27]. In addition, no association was found between ACE genotypes and echocardiographic measurements [28]. However Schunkert and colleagues described an association between DD genotype and electrocardiographic left ventricular hypertrophy in an European population [29].
Serum ACE Activity
The serum ACE activity measured is higher in the HF group with a level of 39.49 UI (26.5-171.3) vs 26.03 UI (12-47.9) in the Non HF group (p=0.08); knowing that the normal range is 17.8 ± 5.4 IU. In our study, we found that the DD genotype had a significantly higher serum ACE activity compared to ID and II (p<0.01). And the highest level of serum ACE activity is noticed in the group of HF who has also hypertension and diabetes, with the highest risk (OR=9.12, CI 95% [1.14-17.4]). Baudin in 2005 confirmed that the elevation of serum ACE activity is a marker of endothelial alteration, increased risk of cardiovascular disease and therapeutic efficacy of treatment with ACE inhibitors [30] and in particular myocardial infarction [31].
Aldosterone Synthase C-344 T Polymorphism
Few studies have described the association between heart failure and C-344 T polymorphism. In our study we found an association between the CYP11B2 polymorphism and the HF group with a significant p=0.025 and an OR=1.267, CI95% [0.478-3.359]. Studies in Caucasian and Japanese patients did not find an association between this variant and idiopathic dilated cardiomyopathy [32]. However, the C allele was associated with increased left ventricular volume in the Japanese patients [33]. But this finding was not confirmed in black South Africans with heart failure [26]. Studies in hypertrophic cardiomyopathy also yielded conflicting results [26,34]. Consequently, there is no firm evidence for the role of this polymorphism in heart failure [1]. In young healthy Finns, the C-344 T variant seems to be associated to the left ventricular hypertrophy [18]. Although one small study confirmed this association [35], other larger studies failed to detect a significant effect of this polymorphism on left ventricular structure [36,37]. Mayosi et al. investigated the contribution of several markers in the CYP11B2 gene, including the C-344 T polymorphism, as individual variants and haplotypes [38]. No significant association was detected with cardiac mass [1].
Renin C-4063 T Polymorphism
Heart Failure (HF) is often complicated by decreased renal blood flow and a subsequent decrease in glomerular filtration rate. In our study we didn’t found any association between the renin C-4063 T polymorphism and HF (p=0,294). Decreased renal function is one of the strongest predictors of mortality in patients with advanced systolic HF [39]. The evidence for a potential effect of renin blockade in diastolic heart failure is rare [10]. Mtiraoui et al, in 2011, evaluated the contribution of the renin -4063C/T promoter variant to the risk for diabetic nephropathy patients. In this study, there was no asso¬ciation between renin -4063T allele or -4063C/ T genotypes and diabetic nephropathy [40]. The RFLP analysis of the study of Shuchi among an Indian Population demonstrated no significant difference between high altitude pulmonary edema and acclimatized control subjects’ groups for the Renin C(–4063)T polymorphism [41]. Data related to Renin C–4063T polymorphism and its association with essential hypertension is also available [42].
Limitations
Some potential limitations should be noted to our study. We do need to measure the physiological values of RAAS and analyze its effect on the association of selected SNPs with the risk of AHF patients among Tunisian population. Second, further investigation in a large sample size on the level of gene-environment interaction might be essential to further verify our results. Moreover, the study has involved a single centre and the limited sample size might have led to low statistical power and trivial difference in between study groups. Hence, a detailed multicentre study on the population from various hospitals has to be performed for better elucidation of the role of RAAS genes polymorphisms on the progression of AHF. Hence population studies linking ACE gene to hypertension, CAD and CKD need to study ACE gene aberrations in the context of ethnicity, age, gender, environmental and geographic factors, and importantly, to the duration of hypertension.
Conclusion
Renin C–4063 T polymorphism showed no association with the risk of developing heart failure in this study. ACE ID and CYP11B2 polymorphisms can serve as a marker for early diagnosis of heart failure as previously reported. In addition, blood ACE activity can be a marker of acute heart failure.
Acknowledgment
We would like to thank the Emergency Department at Fattouma Bourguiba University Hospital in Monastir (Tunisia) for recruiting stroke patients and for their help.
Conflict of Interest
The authors declare no conflicts of interest in preparing this article.
References
- Bleumink G, Schut A, Sturkenboom M, Deckers J, Van Duijn C (2004) Genetic polymorphisms and heart failure. Genetics in Medicine 6(6): 465-474.
- Cuadrado Godia E, Ois A, Roquer J (2010) Heart Failure in Acute Ischemic Stroke. CCR 6(33): 202-213.
- McMurray J, Califf R, Bethel A, Haffner S, Holman R (2012) Comparative Effectiveness of Angiotensin-Converting Enzyme Inhibitors and Angiotensin Receptor Blockers for Hypertension on Clinical End Points: A Cohort Study. The Journal of Clinical Hypertension 14(10): 731-731.
- Hunt S (2005) ACC/AHA 2005 Guideline Update for the Diagnosis and Management of Chronic Heart Failure in the Adult-Summary Article: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Writing Committee to Update the 2001 Guidelines for the Evaluation and Management of Heart Failure): Developed in Collaboration With the American College of Chest Physicians and the International Society for Heart and Lung Transplantation: Endorsed by the Heart Rhythm Society. Circulation 112(12): 1825-1852.
- Peacock WF, Braunwald E, Abraham W, Albert N, Burnett J, et al. (2003) Calpain and caspase-3 inhibitors reduce infarct size and post-ischemic apoptosis in rat heart without modifying contractile recovery. Cell Mol Biol (Noisy-le-grand) 49 Online Pub: OL497-505.
- Braunwald E (2013) Heart Failure. JACC: Heart Failure 1(1): 1-20.
- He J, Ogden L, Bazzano L, Vupputuri S, Loria C (2001) Risk Factors for Congestive Heart Failure in US Men and Women. Arch Intern Med 161(7): 996.
- Nicholls M, Richards A, Agarwal M (1998) The importance of the renin-angiotensin system in cardiovascular disease. J Hum Hypertens 12(5): 295-299.
- Ferrario C (1990) The Renin-Angiotensin System. Journal of Cardiovascular Pharmacology 15: S1-5.
- Schroten N, Gaillard C, Van Veldhuisen D, Szymanski M, Hillege H (2011) New roles for renin and prorenin in heart failure and cardiorenal crosstalk. Heart Failure Reviews 17(2): 191-201.
- Guang C, Phillips R, Jiang B, Milani F (2012) Three key proteases angiotensin-I-converting enzyme (ACE), ACE2 and renin within and beyond the renin-angiotensin system. Archives of Cardiovascular Diseases. 105(6-7): 373-385.
- Higashimori K, Zhao Y, Higaki J, Kamitani A, Katsuya T (1993) Association Analysis of a Polymorphism of the Angiotensin Converting Enzyme Gene with Essential Hypertension in the Japanese Population. Biochemical and Biophysical Research Communications 191(2): 399-404.
- Cambien F, Costerousse O, Tiret L, Poirier O, Lecerf L (1994) Plasma level and gene polymorphism of angiotensin-converting enzyme in relation to myocardial infarction. Circulation 90(2): 669-676.
- Ledru F, Blanchard D, Battaglia S, Jeunemaitre X, Courbon D, et al. (1998) Relation between severity of coronary artery disease, left ventricular function, and myocardial infarction, and influence of the ACE ID gene polymorphism. The American Journal of Cardiology 82(2): 160-165.
- Clyne C, Zhang Y, Slutsker L, Mathis J, White P (1997) Angiotensin II and Potassium Regulate Human CYP11B2 Transcription through Common cis -Elements. Molecular Endocrinology 11(5): 638-649.
- Russo P, Loguercio M, Lauria F, Barba G, Arnout J (2007) Age- and gender-dependent association of the -344C/T polymorphism of CYP11B2 with blood pressure in European populations. J Hum Hypertens 21(4): 333-336.
- Normaznah Y, Azizah MR, Kuak SH, Rosli MA (2015) CYP11B2 gene polymorphism among coronary heart disease patients and blood donors in Malaysia. The Malaysian journal of pathology 37(1): 45-47.
- Kupari M, Hautanen A, Lankinen L, Koskinen P, Virolainen J (1998) Associations Between Human Aldosterone Synthase (CYP11B2) Gene Polymorphisms and Left Ventricular Size, Mass, and Function. Circulation 97(6): 569-575.
- Andersson B, Blange I (1999) Angiotensin-II type 1 receptor gene polymorphism and long-term survival in patients with idiopathic congestive heart failure. European Journal of Heart Failure 1(4): 363-369.
- Raynolds M (1993) Angiotensin-converting enzyme DD genotype in patients with ischaemic or idiopathic dilated cardiomyopathy. The Lancet 342(8879): 073-1075.
- Candy G, Samani N, Norton G, Woodiwiss A, Radevski I (2000) Association analysis of beta2 adrenoceptor polymorphisms with hypertension in a Black African population. Journal of Hypertension 18(2): 167-172.
- Harn HJ, Chang CY, Ho Li (1995) Evidence that polymorphism of the angiotensin I convert enzyme gene may be related to idiopathic dilated cardiomyopathy in the Chinese population. Biochem Mol Biol Int 35(6): 1175-1181.
- Marian A (1993) Angiotensin-converting enzyme polymorphism in hypertrophic cardiomyopathy and sudden cardiac death. The Lancet 342(8879): 1085-1086.
- Bilsel T, Terzi S, Akbulut T, Sayar N, Hobikoglu G (2006) Abnormal Heart Rate Recovery Immediately After Cardiopulmonary Exercise Testing in Heart Failure Patients. International Heart Journal 47(3): 431-440.
- Sanderson J, Young R, Yu C, Chan S, Critchley J (1996) Lack of association between insertion/deletion polymorphism of the angiotensin-converting enzyme gene and end-stage heart failure due to ischemic or idiopathic dilated cardiomyopathy in the chinese. The American Journal of Cardiology 77(11): 1008-1010.
- Tiago A, Badenhorst D, Skudicky D, Woodiwiss A, Candy G (2002) An aldosterone synthase gene variant is associated with improvement in left ventricular ejection fraction in dilated cardiomyopathy. Cardiovascular Research 54(3): 584-589.
- Candy G, Skudicky D, Mueller U, Woodiwiss A, Sliwa K (1999) Association of left ventricular systolic performance and cavity size with angiotensin-converting enzyme genotype in idiopathic dilated cardiomyopathy. The American Journal of Cardiology 83(5): 740-744.
- Yamada Y (1997) Lack of Association of Polymorphisms of the Angiotensin Converting Enzyme and Angiotensinogen Genes With Nonfamilial Hypertrophic or Dilated Cardiomyopathy. American Journal of Hypertension 10(8): 921-928.
- Schunkert H, Hense H, Holmer S, Stender M, Perz S (1994) Association between a Deletion Polymorphism of the Angiotensin-Converting-Enzyme Gene and Left Ventricular Hypertrophy. New England Journal of Medicine 330(23): 1634-1638.
- Baudin B (2005) Polymorphism in angiotensin II receptor genes and hypertension. Experimental Physiology 90(33): 277-282.
- Bas M, Hoffmann T, Tiemann B, Dao V, Bantis C, et al. (2010) Potential genetic risk factors in angiotensin-converting enzyme-inhibitor-induced angio-oedema. British Journal of Clinical Pharmacology 69(2): 179-186.
- Tiret L, Mallet C, Poirier O, Nicaud V, Millaire A (2000) Lack of association between polymorphisms of eight candidate genes and idiopathic dilated cardiomyopathy. Journal of the American College of Cardiology 35(1): 29-35.
- Takai E (2002) Association between aldosterone synthase (CYP11B2) gene polymorphism and left ventricular volume in patients with dilated cardiomyopathy. Heart 88(6): 649-650.
- Patel R, Lim D, Reddy D, Nagueh S, Lutucuta S, et al. (2000) Variants of Trophic Factors and Expression of Cardiac Hypertrophy in Patients with Hypertrophic Cardiomyopathy. Journal of Molecular and Cellular Cardiology 32(12): 2369-2377.
- Delles C, Erdmann J, Jacobi J, Hilgers K, Fleck E (2001) Aldosterone synthase (CYP11B2)-344 C/T polymorphism is associated with left ventricular structure in human arterial hypertension. Journal of the American College of Cardiology 37(3): 878-884.
- Schunkert H, Hengstenberg C, Holmer S, Broeckel U, Luchner A, et al. (1999) Lack of Association Between a Polymorphism of the Aldosterone Synthase Gene and Left Ventricular Structure. Circulation 99(17): 2255-2260.
- Hengstenberg C, Holmer S, Mayer B, Lowel H, Engel S (2000) Evaluation of the Aldosterone Synthase (CYP11B2) Gene Polymorphism in Patients With Myocardial Infarction. Hypertension 35(3): 704-709.
- Mayosi B, Keavney B, Watkins H, Farrall M (2003) Measured haplotype analysis of the aldosterone synthase gene and heart size. Eur J Hum Genet 11(5): 395-401.
- Grigorian Shamagian L, Roman A, Ramos P, Veloso P, Bandin Dieguez M (2006) Angiotensin-Converting Enzyme Inhibitors Prescription Is Associated With Longer Survival Among Patients Hospitalized for Congestive Heart Failure Who Have Preserved Systolic Function: A Long-Term Follow-Up Study. Journal of Cardiac Failure 12(2): 128-133.
- Mtiraoui N, Ezzidi I, Turki A, Chaieb M, Mahjoub T (2011) Renin-angiotensin-aldosterone system genotypes and haplotypes affect the susceptibility to nephropathy in type 2 diabetes patients. Journal of the Renin-Angiotensin-Aldosterone System 12(4): 572-580.
- Bhagi S, Srivastava S, Tomar A, Bala Singh S, Sarkar S (2015) Positive association of D Allele of ACE Gene With High Altitude Pulmonary Edema in Indian Population. Wilderness & Environmental Medicine 26(2): 124-132.
- Chiang F, Lai Z, Chern T, Tseng C, Hsu K (1997) Lack of Association between Angiotensin-converting Enzyme Gene Polymorphism and Coronary Heart Disease in a Chinese Population. Jpn Heart J 38(2): 227-236.

 Research Article
Research Article