Abstract
Viral diseases have greatly affected human lives and civilizations from time immemorial. The Newcastle disease virus (NDV) is causing lethal and highly contagious diseases in birds especially chickens and having a negative impact worldwide on the poultry industry. Although this virus has been effectively vaccinated, it is still prevalent in Pakistan and disease-related outbreaks have led to serious losses regarding mortality and loss in meat and egg production for the poultry industry in the country. Different genes of this negative single-stranded RNA virus have been sequenced, cloned, and expressed in plasmid-vectors in various organisms. There are different strategies to produce vaccines of NDV and plant-based vaccine (PBV) is one of them. Fusion (F) gene has the most virulence impact among the all-other genes of NDV. In this study, the epitope gene of NDV was cloned into the plant expression vector. Agrobacteriummediated transformation of F gene in Nicotiana benthamiana was performed through the leaf disc method. The epitope was checked by amplification using specific primers through polymerase chain reaction (PCR). Real-time polymerase chain reaction (RT-PCR) was carried out to analyze the expression of the transgene in transgenic N. benthamiana plants. Through this research, we might be able to fight any pandemic disease within few months instead of producing a vaccine in several years in the future.
Keywords: Poultry; Newcastle Disease Virus; Agrobacterium-Mediated Transformation; Plant-Based Vaccine
Introduction
Viruses are dominating on the earth as a competitor of humanity as a parasite and replicating their genetic material in their hosts [1]. Healthy food of a high quality at reasonable commercial prices is the priority of the consumers. There are different sources of meat proteins in the world, but chicken meat is the cheapest among them. Fast food restaurants like Fri Chicks, McDonald’s, Forks & Knives, and Kentucky Fried Chicken (KFC) in Pakistan have significantly increased their chicken consumption during the last years. It is estimated that the total annual revenue of Pakistan’s poultry industry is Rs. 1168 billion. However, the industry often has out breaks of several disorders, like bacterial bronchitis, avian influenza, chicken-like anemia, and NDV.
Newcastle disease (ND) is caused by an unsegmented singlestranded RNA virus which is enveloped in a protein coat with an adverse polarity. This virus consists of six genes, with their corresponding six structural proteins: large polymerase (L), matrix (M), nucleoprotein (NP), phosphoprotein (P), fusion (F), and hemagglutinin-neuraminidase (HN). The editing of P-protein RNA produces two additional proteins, V and W. HN and F encode for glycoproteins that bind and fuse the virus to the host cells and cause NDV infection. The primary protective factor of ND vaccines is HN and F proteins [2]. F glycoprotein is the primary virulence factor of the NDV [3]. The precursor F0 is composed of 553 amino acids and has a 55 kDa molecular mass [4,5]. The F glycoprotein has five or six prospective sites. There are probably 20 residues of fusion peptide conserved in the hydrophobic zone of F1 fragment amino terminus. Viral and host cell membrane connection is regulated through this region [6].
Natural pathogens such as Agrobacterium tumefaciens are utilized to integrate antigenic DNA stably in the plant’s nucleus. The inherent capacity of pathogens to infect and transfer their virulent genes to the host cell nucleus benefits transformation [7]. The transgene can be integrated into the nuclear or plastid genome either through transient or stable transformation [8]. Stable transformation is achieved through the permanent integration of the transgene. Many plants are restricted to agrobacteriummediated transformation [9]. Many plants have been used for the expression of transgenes but Nicotiana benthamiana has significant importance for the expression of therapeutic recombinant proteins [10]. N. benthamiana is an Australian native with many features, including the rapid growth and natural capacity for biopharmaceuticals in heterologous gene sequences [11]. The viral or binary vector system is often involved in the vacuum infiltration of leaf tissue with agrobacterium containing transgene for a protein of interest [12,13]. The availability of recombinant protein in the plant is based on an extensive amount of protein in one gram of leaf tissue. Plant leaf biomass is counted before agro-infiltration and after it [14,15]. Recombinant proteins in N. benthamiana and original plant proteins have maximum structural resemblance and stability in plant cells environment [16,17].
In 1992, the concept of the production of vaccines in transgenic plants was introduced. In 1995, a biopharmaceutical company Pros gene from Texas (USA) carried out clinical studies on pigs, which were supplied with an edible maize vaccine, and they were found to be protected against TGEV. Patents were also obtained for this and other edible transgenic crop vaccines. Such vaccines provide technology, particularly in the developing world, for the production and supply of cheap vaccines, wherever they succeed. Plant-based platforms are one of the preferred expression systems active for recombinant vaccine assembly [18].
Materials and Methods
Vector Designing
The binary vector pHNF was designed by the ligating pBR 322 ori with pVS1 oriV, pVS1 RepA, pVS1 StaA, and kanamycin sequence (https://en.vectorbuilder.com). The ORF of 376 bp was designed along with CaMV 35S_short and Nos pA within the left and right border of the construct (Figure 1), there were four tandem repeats of HN epitope (amino acid encoding 346–353) with the length of 96 bp and three F epitope (amino acid encoding 65–81) repeats of the tandem with the length of 153 bp in an ORF [19].
Figure 1: NDV epitope gene cassette: Gene construct including F and HN epitopes together with needed sequences for expression in N. benthamiana
Gen Script’s Optimum Gene TM optimization tool (https:// www.genscript.com/codon-opt.html) was used to analyze the NDV sequence of the genes (HN and F) to guarantee a high level of expression. At the upstream of the ORF, initiation codon (AUG), followed by histidine tag (18 bp) and omega sequence as a ribosome binding (67 bp) were attached. A 3′′ end was included before the ending of the codon with the endoplasmic reticulum signal (SEKDEL). Lastly, at five and three ends, two restriction sites were added, namely BamHI and SacI. The sequence was analyzed to find the open reading frames (ORF) with the ORF Finder tool (https:// www.ncbi.nlm.nih.gov/orffinder/).
SWISS-MODEL tool (https://swissmodel.expasy.org/) was used to predict the 3D structure of the optimized codons’ protein.
Vector Cloning and Confirmation
The vector pHNF was cloned into E. coli through heat shocking of 90 seconds in the water bath at 42ᵒC. The plasmid was isolated through Thermo Scientific Gene JET Plasmid Miniprep kit (www. thermoscientific.com/onebio) from 5ml culture of a transformed E. coli single-cell colony. The vector was confirmed by XhoI restriction digestion. The reaction was incubated for 3 hours at 37ᵒC and run on 1% agarose gel along with Fastlane 1Kb DNA ladder.
Transformation of NDV Epitope Gene into N. benthamiana
The sterilized leaf discs of N. benthamiana were dipped in LB4404 culture containing acetosyringone for 30 minutes to follow the Agrobacterium-mediated leaf disc transformation method. The leaf discs were kept on MS co-culture media in dark for 2 days after treating with LB4404 culture. The leaf discs were washed with cefotaxime (250mg/L) and kept on kanamycin selection MS shoot regeneration media after 2 days of dark incubation. The emerging shoots were excised and shifted into kanamycin selection MS root regeneration media after 4 weeks. Harding of the transgenic regenerated plants was done in 3-4 weeks. T0 seeds were collected for further generation.
Gene Expression Analysis
Genomic DNA was extracted from young leaves of transgenic seedlings and wild-type N. benthamiana by the CTAB method to determine the presence of foreign genes in the genome of the regenerated kanamycin-resistant seedlings. PCR was carried out by using specific primers (5′ACTATTACAATTACATGCATCA3′ and 5′GAGTTCATCTTTCAAAGTG3′). The 35 cycles for the PCR were 94ᵒC, 60 s for 55ᵒC, and 60 s for 72ᵒC, followed by a final extension of 72ᵒC, which was completed in 10 minutes. As a control, genomic DNA of N. benthamiana was used. 1.5% agarose gel was used to separate the PCR product along with the omega BIO-TEK 100bp DNA ladder.
Total RNA was extracted using a commercial kit of Triazole. Following DNase treatment, cDNA was synthesized by using the cDNA synthesis kit. cDNA was synthesized by using oligo(dT) as reverse primer (5′-GCGGGATCCTTTTTTTTTTTTTTTTTT-3′). RT-PCR was carried out with specific primers for verification of transgene expression in transgenic seedlings of N. benthamiana (forward: 5′ACTATTACAATTACATGCATCA3′; reverse: 5′GAGTTCATCTTTCAAAGTG3′). PCR cycles included 1minute denaturation, 55ᵒC for 45 s, and 72ᵒC for 45 s at 94ᵒC. cDNA of wild plant type was used as controls. 2% agarose gel was used to separate the RT-PCR product along with omega BIO-TEK 100bp DNA ladder.
Result
Vector Designing
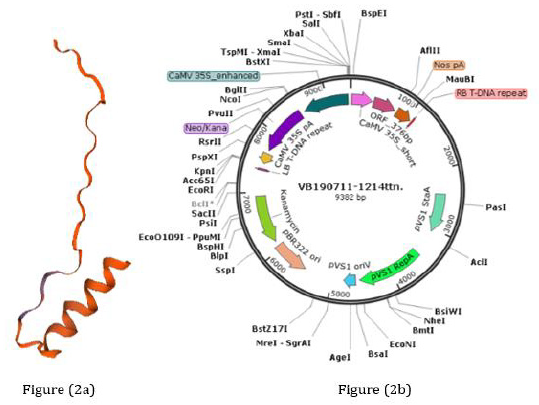
Codons of four tandem repeats of HN 96bp and three tandem repeats of F 156bp were optimized by Gen Script’s Optimum Gene TM optimization tool (http:/www.genscript.com/codon-opt.html) for the expression analysis. 3D structure of predicted proteins by SWISS-MODEL tool (https://swissmodel.expasy.org/) from these codons resembled NDV epitope protein (Figure 2a). The binary plant expression vector was designed with a 35S promotor and NospA terminator along with kanamycin antibiotics selection (Figure 2b).
Figure 2: Predicted protein and vector pHNF:
(a) Predicted 3D structure of protein produce by tandem repeats of HN and F genes of NDV and resembled with NDV epitope protein through SWISS-MODEL tool (https://swissmodel.expasy.org/).
(b) pHNF plant expression vector with tandem repeats of HN and F genes of NDV, synthesized by Vector Builders (https:// en.vectorbuilder.com).
Vector Cloning and Confirmation
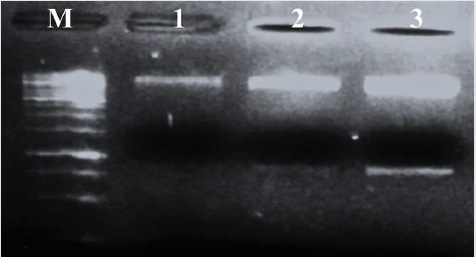
The isolated plasmid was digested with restriction enzyme XhoI and run on 1% gel (Figure 3). This restriction sequence was placed in pHNF at two sites during vector designing. Comparison of digested fragments (8491bp and 891bp) with Fastlane 1Kb DNA ladder confirmed that the isolated plasmid was pHNF and it was successfully cloned in E. coli.
Figure 3: Restriction digestion: digestion with XhoI at specific double cutter sites in pHNF. Lanes 1, 2, and 3 contain two bands of 8491 bp and 891 bp, and lane M contains Fastlane 1Kb DNA Ladder. Band intensity in lane 1, 2, and 3 shows the quantity of the plasmid.
Transformation of NDV Epitope Gene into N. benthamiana
Leaves of N. benthamiana plants were transformed using A. tumefacien strain LBA4404 with pHNF through the leaf disc method. Almost two hundred pre-cultured leaf discs (Figure 4a) of N. benthamiana were co-cultured (Figure 4b). A thin whitish film of the bacterial growth was seen on the periphery of infected discs which became more visible after 72 hours and also covered the surface of discs after one day of co-culture. It was observed that due to kanamycin selection pressure only the transformed cells were developed into shoots (Figure 5) after 28 days. The shoots were shifted on selection rooting media to develop the shoots into the whole plant (Figure 6a-6d).
Figure 4: Leaf discs on MS media:
(a) Leaf discs of N. benthamiana on pre-culture media.
(b) Leaf discs of N. benthamiana on co-culture media after treating with Agrobacteria containing pHNF.
Figure 5: Shoots regeneration from the leaf disc. Shoots regenerating from the leaf discs of N. benthamiana on selection media with shooting plant growth regulators.
Figure 6: Plant development from the shoot:(a) Shifting of shoots (regenerated from leaf disc of N. benthamiana on selection media after treating with Agrobacteria containing pHNF) into selection rooting media (b) and (c) Growth and development of shoots (regenerated from leaf disc of N. benthamiana on selection media after treating with Agrobacteria containing pHNF) on selection rooting media (d) Shoots developing into the whole plant on selection media with rooting plant growth regulators.
Gene Expression Analysis
The transgene was determined by PCR amplification with specific forward and reverse primers from transgenic plants. The 307bp fragments of the transgene were amplified and represented the presence of the transgene in transgenic N. benthamiana (Figure 7). The amplification of desired size products confirmed the successful transformation of the transgene, while no amplification was detected in the untransformed negative control (N. benthamiana). RT-PCR gave the same results according to the PCR but the band intensity was different for putative transgenic plants (Figure 8). Different intensity levels indicated the amount of mRNA transcription in cells of each transgenic plant.
Figure 7: Gel picture of PCR: PCR amplification of NDV epitope gene from transgenic N. benthamiana seedlings. Lane 1 to 9 contains 307bp amplified product of NDV epitope gene. While Lane M contains omega BIO-TEK 100bp ladder and Lane 10 represents control (wild N. benthamiana). Band intensity in lanes shows the quantity of the PCR product.
Figure 8: Gel picture of RT-PCR: RT-PCR amplification of NDV epitope gene from transgenic N. benthamiana seedlings. Lane 1, 2, 3, and 4 contains 307bp amplified product of the NDV epitope gene. While Lane M contains omega BIO-TEK 100bp ladder and Lane 5 represent control (wild N. benthamiana). Band intensity in lanes show the quantity of the RT-PCR products.
Discussion
Tobacco is the most preferred model in bioreactor plant studies for the expression, due to its easy transformation and its rapid regeneration, of foreign protein [20,21]. Therefore, in the present study, N. benthamiana plants were used as a model for genetic transformation and the evaluation of NDV epitope production. A synthetic gene with 4 tandem repeats of HN epitopes and 3F NDV epitopes has been prepared in the current research. Based on studies, the optimization of a codon can increase protein quality and quantity. The gene construction has been optimized based on the preferred codon. 5′ omega (TMV), which is considered to be a translation enhancement element, has been added to 5′UTR. In terms of the effect of SEKDEL on gene expression improvement. SEKDEL had been attached to 3′ before the stop codon. To identify and isolate target protein by anti-His sequence, a Histidine tag was added to amino gene terminals [22].
Confirmation of the vector is necessary before using it for the transformation. Vector pHNF was cloned in E. coli to increase the quantity of the vector for the experiments. The plasmid was isolated from single colony culture to avoid contamination. The same size of the bands showed that extracted plasmids were the same and there was no contamination during culture growth and plasmid isolation. The extracted plasmid samples were digested with restriction enzyme XhoI. The specific sequence for the XhoI enzyme was placed at two sites in the vector pHNF. The band size of the pHNF fragments was confirmed through Snapgene viewer software (http://www.snapgene.com/) by finding XhoI double cutter sites in pHNF. Approximately, 8491bp and 891bp fragments were obtained which confirmed that cloned vector was pHNF. Confirmation of the vector through restriction digestion has been followed by many scientists, Chaudhury et al. [23] is one of them.
Plants of N. benthamiana were transformed through LBA4404 A. tumefacien strain with plasmid pHNF. Genetic transformation by the Agrobacterium strain has various advantages, including the large piece of DNA transformation, increased transformation efficiencies, low copy number addition, and minimum reorganization of transferred DNA [24,25]. Genomic DNA and RNA were isolated at 3-4 leaf stage and subjected to PCR and RT-PCR amplification, respectively. Total DNA was extracted to screen for the foreign gene using PCR after germination of transgenic seedlings. PCR primers have been designed to match the ORF of pHNF. Approximately, 307bp NDV epitope gene fragment was obtained from 9 putative N. benthamiana plants, PCR confirmed the presence of transgene (Figure 7) whereas no band was observed for non-transgenic control plants. Band intensity was almost the same for all the tested plants which were indicated that the integration was the same and successful. The analysis of gene integration in transgene through PCR is a general practice that has been followed by several scientists [26-28]. RNA expression in four plants using RT-PCR was semi-quantitatively measured. RT-PCR gave the same results according to the PCR, but the band intensity was different for putative transgenic plants. Different intensity levels indicated the amount of mRNA transcription in cells of each transgenic plant [29-31].
Conclusion
The plant-based vaccine of NDV was first time produced in Pakistan and it was observed that many other diseases have threats to the poultry industry. Expression of NDV epitope gene in Nicotiana benthamiana justified that other antigenic ORFs could be expressed in plants and used as a vaccine. Further studies are also required to evaluate the plant based NDV vaccine effectiveness and economic values in comparison with other commercial vaccines. There is also a need to develop an edible plant-based vaccine to ease the vaccine administration.
Acknowledgement
We acknowledge Dr. Sultan Habibullah Khan and Dr. Aftab Ahmed, Center for Advanced Studies in Agriculture and Food Security (CAS-AFS), University of Agriculture, Faisalabad, Pakistan for providing technical support and guidance during this research.
Funding
The project was completed using personal funds of PI (ZK).
Conflicts of interest/Competing Interests
Authors declare that they have no conflict of interest.
Authors’ Contributions
ZK gave idea and funds and supervised the research experiments. MS conducted research experiments and wrote the manuscript under the supervision of ZK. ZA help MS in the research experiments. ZK, UW and GA reviewed final draft.
Ethics Approval
This chapter does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
References
- Lederburg J (1994) Emerging infections: private concerns and public responses ASM News 60: 233.
- Xiao S, Baibaswata Nayak, Arthur Samuel, Anandan Paldurai, Mallikarjuna Kanabagattebasavarajappa, et al. (2012) Generation by reverse genetics of an effective, stable, live-attenuated Newcastle disease virus vaccine based on a currently circulating, highly virulent Indonesian sstrain. PloS one 7: e52751.
- Römer-Oberdörfer A, Werner O, Veits J, Mebatsion T, Mettenleiter TC (2003) Contribution of the length of the HN protein and the sequence of the F protein cleavage site to Newcastle disease virus pathogenicity. Journal of General Virology 84: 3121-3129.
- Chambers P, Millar NS, Emmerson PT (1986) Nucleotide sequence of the gene encoding the fusion glycoprotein of Newcastle disease virus. Journal of general virology 67:2685-2694.
- Salih O, Omar A, Ali A, Yusoff K (2000) Nucleotide sequence analysis of the F protein gene of a Malaysian velogenic NDV strain AF2240. Journal of Molecular Biology, Biochemistry & Biophysics 4: 51-57.
- Lamb RA (2001) Paramyxoviridae: the viruses and their replication Fields virology.
- Jacob SS, Cherian S, Sumithra T, Raina O, Sankar M (2013) Edible vaccines against veterinary parasitic diseases-current status and future prospects. Vaccine 31: 1879-1885.
- Altpeter F, Niranjan Baisakh, Roger Beachy, Ralph Bock, Teresa Capell, et al. (2005) Particle bombardment and the genetic enhancement of crops: myths and realities. Molecular Breeding 15: 305-327.
- Ma H, Chen G (2005) Gene transfer technique Nature and Science 3: 25-31.
- Bally J, Hyungtaek Jung, Cara Mortimer, Fatima Naim, Joshua G Philips, et al. (2018) The rise and rise of Nicotiana benthamiana: a plant for all reasons. Annual review of phytopathology 56: 405-426.
- Lomonossoff GP, D’Aoust MA (2016) Plant-produced biopharmaceuticals: a case of technical developments driving clinical deployment. Science 353: 1237-1240.
- Leuzinger K, Dent M, Hurtado J, Stahnke J, Lai H, Zhou X, Chen Q (2013) Efficient agroinfiltration of plants for high-level transient expression of recombinant proteins. JoVE (Journal of Visualized Experiments): e50521.
- Norkunas K, Harding R, Dale J, Dugdale B (2018) Improving agroinfiltration-based transient gene expression in Nicotiana benthamiana. Plant methods 14: 1-14.
- Fujiuchi N, Matoba N, Matsuda R (2016) Environment control to improve recombinant protein yields in plants based on Agrobacterium-mediated transient gene expression. Frontiers in bioengineering and biotechnology 4: 23.
- Shang L, Gaudreau L, Martel M, Michaud D, Pepin S, et al. (2018) Effects of CO 2 enrichment, LED inter-lighting, and high plant density on growth of Nicotiana benthamiana used as a host to express influenza virus hemagglutinin H1. Horticulture, Environment, and Biotechnology 59: 637-648.
- Faye L, Boulaflous A, Benchabane M, Gomord V, Michaud D (2005) Protein modifications in the plant secretory pathway: current status and practical implications in molecular pharming. Vaccine 23: 1770-1778.
- Gomord V, Fitchette AC, Menu‐Bouaouiche L, Saint‐Jore‐Dupas C, Plasson C, et al. (2010) Plant‐specific glycosylation patterns in the context of therapeutic protein production. Plant biotechnology journal 8: 564-587.
- Sala F, Rigano MM, Barbante A, Basso B, Walmsley AM, et al. (2003) Vaccine antigen production in transgenic plants: strategies, gene constructs and perspectives. Vaccine 21: 803-808.
- Ghaffar Shahriari A, Bagheri A, Bassami MR, Malekzadeh-Shafaroudi S, Afsharifar A, Niazi A (2016) Expression of Hemagglutinin-Neuraminidase and fusion epitopes of Newcastle Disease Virus in transgenic tobacco Electronic. Journal of Biotechnology 19: 38-43.
- Jutras PV, Carla Marusic, Chiara Lonoce, Carole Deflers, Marie-Claire Goulet, et al. (2016) An Accessory Protease Inhibitor to Increase the Yield and Quality of a Tumour-Targeting mAb in Nicotiana benthamiana Leaves. PloS one 11: e0167086.
- Robert S, Khalf M, Goulet M-C, D’Aoust M-A, Sainsbury F, et al. (2013) Protection of Recombinant Mammalian Antibodies from Development-Dependent Proteolysis in Leaves of Nicotiana benthamiana. PLOS ONE 8: e70203.
- Shahriari AG, Bagheri A, Bassami MR, Malekzadeh Shafaroudi S, Afsharifar AR (2015) Cloning and expression of fusion (F) and haemagglutinin-neuraminidase (HN) epitopes in hairy roots of tobacco (Nicotiana tabaccum) as a step toward developing a candidate recombinant vaccine against Newcastle disease. Journal of Cell and Molecular Research 7: 11-18.
- Chaudhury A, Dalal AD, Sheoran NT (2019) Isolation, cloning and expression of CCA1 gene in transgenic progeny plants of Japonica rice exhibiting altered morphological traits. PloS one 14: e0220140.
- Lin J, Bo Zhou, Yuanzhu Yang, Jin Mei, Xiaoying Zhao, et al. (2009) Piercing and vacuum infiltration of the mature embryo: a simplified method for Agrobacterium-mediated transformation of indica rice. Plant cell reports 28: 1065-1074.
- Xiao F, Giavalisco P, Martin GB (2007) Pseudomonas syringae type III effector AvrPtoB is phosphorylated in plant cells on serine 258, promoting its virulence activity. Journal of Biological Chemistry 282: 30737-30744.
- Bazzini A, Asurmendi S, Hopp H, Beachy R (2006) Tobacco mosaic virus (TMV) and potato virus X (PVX) coat proteins confer heterologous interference to PVX and TMV infection, respectively. Journal of general virology 87:1005-1012.
- Missiou A, Kalantidis K, Boutla A, Tzortzakaki S, Tabler M, et al. (2004) Generation of transgenic potato plants highly resistant to potato virus Y (PVY) through RNA silencing. Molecular Breeding 14:185-197.
- Thomas CL, Leh V, Lederer C, Maule AJ (2003) Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306: 33-41.
- Ghosh S, Nagi L, Ganapathi T, Khurana SP, Bapat V (2002) Cloning and sequencing of potato virus Y coat protein gene from an Indian isolate and development of transgenic tobacco for PVY resistance. Current Science: 855-859.
- Jan FJ, Pang SZ, Fagoaga C, Gonsalves D (1999) Turnip mosaic potyvirus resistance in Nicotiana benthamiana derived by post‐transcriptional gene silencing. Transgenic research 8: 203-213.
- Mäki-Valkama T, Valkonen JP, Kreuze JF, Pehu E (2000) Transgenic resistance to PVYO associated with post-transcriptional silencing of P1 transgene is overcome by PVYN strains that carry highly homologous P1 sequences and recover transgene expression at infection. Molecular Plant-Microbe Interactions 13: 366-373.

 Research Article
Research Article