Abstract
Bladder cancer is the 10th most common cancer worldwide, with significant morbidity and mortality. Its early diagnosis is crucial and there is an urgent need for the development of non-invasive and specific biomarkers to detect bladder cancers at an early stage. Epigenetic alterations are innovative cancer biomarkers that show great potential for assay development to assist in patient management because of their stability and accessibility in body fluids. Recent studies demonstrated that microRNAs (miRNAs) are molecules involved in the epigenetic events emerging as diagnostic biomarkers and either their downregulation or upregulation occurs through epigenetic changes. Urological neoplasms (prostate, bladder, kidney), lung, breast and colorectal cancers are the most common and despite major advances in their characterization, this has not yet translated into biomarkers suitable for clinical practice. However, large multicenter validation studies are required to foster translation to the clinics. Herein we review the most promising epigenetic, diagnostic and predictive biomarkers in different types of biological sample related to patients harbouring bladder cancer.
Keywords:Bladder Cancer; MiRNAs; Epigenetic Modifications; Biomarkers; Biological Sample; Diagnosis
Abbreviations:Ago2: Argonaute Protein-2; Bca: Bladder Cancer; CIS: Carcinoma In Situ; CK20: Citokeratin 20; CTCs: Circulating Tumor Cells; ESCRT: Endosomal Sorting Complex Required for Transport; Evs: extracellular vesicle; FFPE: Formalin-Fixed Paraffin- Embedded; FSCN1: Ascin Actin-Bundling Protein 1; hnRNPA-1: Heterogeneous Nuclear Ribonucleoprotein A1; hnRNPs: Sumoylated Heterogeneous Nuclear Ribonucleoproteins; hTERT: Human Telomerase Reverse Transcriptase; hTR: mRNA- Human Telomerase RNA; MBV: Multivesicular Body; MIBC: Muscle Invasive Bladder Cancer; miRISC: MiRNA Induced Silencing Complex; miRNA: MicroRNA; MSCs: Mesenchymal Stem Cells; NF-κB: Nuclear Factor k-Light-Chain-ENHANCER OF ACTIVATED B CELLS; NMIBC: Non Muscle Invasive Bladder Cancer; nSMase-2: Neural Sphingomyelinase 2; Pre-miRNA: Precursor miRNA; pri-MiRNA: Primary MiRNA; PTEN: Phosphatase And Tensin Homolog; SOX9- SRY: Box Transcription Factor 9; Ta: Low Grade Tumor; tiRNA: tRNA Halves; TP53: Tumor Protein p53; TRAP assay: Telomeric Repeat Amplification Protocol; TRBP: Transactivation Response Element RNA-Binding Protein; tRFs: tRNA-Derived Fragments; VUC: Voided Urine Cytology; WBCs: White Blood Cells; ZEB: Zinc Finger E-Box Binding Homeobox
Mini Review
Among all malignancies, bladder cancer (BCa) deserves particular attention, since is the most common malignancy of the urinary tract in the world. Approximately 70-75% are non-muscle invasive bladder cancer (NMIBC) with an incidence of 430.000 cases newly diagnosed every year causing over 165.000 cancerrelated deaths mortality [1]. It affects the superficial mucosa and submucosa with reduced tendency to cancer progression and high long-term survival. In 25-30% of cases, the disease invades the muscle layer of the bladder wall and it is defined muscleinvasive bladder cancer (MIBC) with a high risk of mortality [1]. Current diagnostic and prognostic strategies, such as tumor grade, stage, size and number of foci, are partially useful for clinical decision-making, in fact, the early diagnosis, disease recurrence and progression are the difficulties for clinicians when caring for patients with BCa. Cystoscopy and voided urine cytology (VUC) are specific diagnostic tools for BCa [2]. Although considered the gold standard, cystoscopy is both invasive and expensive for the patient and can miss certain lesions, in particular small areas of carcinoma in situ (CIS). VUC has a median sensitivity of only 35%, and a median specificity of 94% [3] although it is useful for detecting high‐grade tumors and CIS. However, because of its low sensitivity in lowgrade cancers, it has limited clinical relevance. The limitations of VUC and the invasiveness of cystoscopy have generated interest in other non-invasive tools.
Molecular Markers in BCa
As in other forms of cancer, BCa also shows chromosomal
anomalies. With fluorescence in situ hybridization (FISH)
techniques, chromosomal anomalies can be detected in exfoliated
bladder cells. Several studies reported low sensitivity of FISH to
detect low-grade (36%–57%) and low-stage (62%-65%) tumors,
while has high sensitivity for both high-grade and high-stage
tumors (83%-97%) [4,5]. The detection of CIS is close to 100% and
its hight specificity (89%-96%) is comparable to cytology [6]. An
important multicentre study conducted on 1182 subjects affected
by painless hematuria define the ability of immunocytology to
predict the likelihood of BCa [7]. Immunocytology is based on the
visualization of tumor-associated antigens in BCa using monoclonal
antibodies and results showed that sensitivity, specificity, positive
and negative predictive value of immunocytology for predicting BCa
were 82.4%, 86.6%, 61.6%, and 95.0%, respectively. These data
confirm immunocytology as a strong BCa predictor, outperforming
cytology and helping in the clinical decision-making process.
From voided urine, Eissa and colleagues, analyzed relative
telomerase activity by telomeric repeat amplification protocol
(TRAP assay), human telomerase RNA (hTR-mRNA) by RTPCR
and human telomerase reverse transcriptase (hTERT) by
RT–PCR. Overall the sensitivity of human telomerase reverse
transcriptase for detecting BCa was the highest compared to that
of human telomerase RNA, relative telomerase activity and urine
cytology (96%, 92%, 75% and 75%, respectively). Specificity was
96%, 89%, 92% and 94%, respectively. hTERT followed by hTR
provided sensitivity of 100% and specificity of 85% and thus an
improvement in diagnosis of BCa [8]. In a systematic review, Glas
showed that telomerase had the best sensitivity (75%) compared
to the other markers, including cytology [9]. Among all candidate
biomarkers for BCa, cytokeratin 20 (CK20), a low molecular- weight
protein, measured on tissue from human primary and metastatic
carcinomas after surgical removal, is considered a marker of
urothelial differentiation [10] and several studies explored its role
at the mRNA and protein level [11,12]. A recent study, highlighted
the use of CK20 and p53 immunohistochemistry in bladder
specimens with borderline or suspicious features for urothelial CIS
and correlate them with subsequent or prior cancer diagnoses [13]
Significant difference in CK20 expression was found between low
and high grade urothelial carcinomas; a diffuse positive expression
in 68.8% and 40.6% of high and low grade tumors, was shown,
respectively. Strong positive expression of p53 was seen in 72.9%
of cases of high grade, while only 36.2% cases of low-grade tumors
revealed strong expression. Moreover, they verified the combined
expression of P53 and CK20 in 40% of high grade and 22% of lowgrade
carcinomas [13].
Implication of MiRNAs During Carcinogenesis
The recent literature highlights the importance of specific
molecules that regulate protein-coding genes post transcriptionally
in higher eukaryotes. Of particular interest in this field of
molecular research are miRNAs [14], a class of non-coding RNAs
of 19 to 24 nucleotides that regulate gene expression through posttranscriptional
pathways, RNA interference and gene silencing
[15]. Similar to epigenetic mechanisms, miRNAs appear to be
important in the onset and development of several diseases. In
humans, miRNAs involvement in cancer is spurring the interest
of scientist [16], since they contain a great amount of information
about the regulatory mechanisms of the cells. In attempts to
classify various forms of cancer with expression profiles, miRNAs
have been shown to harbor more information about disease states
than mRNA profiles do [17]. MiRNAs are transcribed using RNA
polymerase II into an up to 3000 nucleotide long primary miRNA
(pri-miRNA), which is processed by Drosha to the precursor miRNA
(pre-miRNA); a single pri-miRNA may contain multiple miRNA
precursors [18]. The RNAse III enzyme Dicer cuts the pre-miRNAs
into 17-24 nucleotide long miRNA–miRNA duplex. The interaction
of Dicer with transactivation response element RNA-binding
protein (TRBP) leads to the formation of mature, single-stranded
miRNA molecules that can regulate approximately one-third of the
human genome post-transcriptionally, allowing for the epigenetic
modulation of important cellular processes like apoptosis or
proliferation [19]. The different expression of these molecules is easily measured, and the miRNA expression profiles were shown to
be altered in tumor cells compared with normal tissues [20].
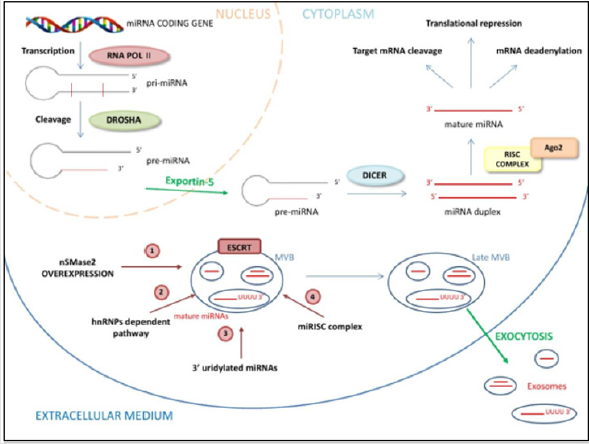
Recently, miRNAs have been identified in exosomes where their
biogenesis, release and uptake may involve the endosomal-sorting
complex required for transport (ESCRT). This machinery is made
up of cytosolic protein complexes, known as ESCRT-0, ESCRT-I,
ESCRT-II, and ESCRT-III, together with a number of accessory
proteins, playing a pivotal role in cellular processes including
multivesicular body biogenesis of exosome, cellular abscission and
viral budding [21]. Exosomes are 40–100 nm nano-sized vesicles
that are released from many cell types into the extracellular space.
Such vesicles are widely distributed in various body fluids [22].
Exosomal miRNAs play an important role in disease progression by
stimulating angiogenesis and facilitate metastasis in cancers. Based
on current research, there are four potential modes for sorting
of miRNAs into exosomes, although the underlying mechanisms
remain largely unclear. These include:
a. The neural sphingomyelinase 2 (nSMase2)-dependent
pathway where the overexpression of nSMase2 increased the
number of exosomal miRNAs, and conversely inhibition of
nSMase2 expression reduced the number of exosomal miRNAs
[23];
b. The miRNA motif and sumoylated heterogeneous nuclear
ribonucleoproteins (hnRNPs)-dependent pathway that causes
specific miRNAs to be packed into exosomes [24];
c. The 3’ end of the miRNA sequence-dependent pathway.
Koppers-Lalic and colleagues discovered that the 3’ ends of
uridylated endogenous miRNAs were mainly presented in
exosomes derived from B cells or urine, whereas the 3’ ends
of adenylated endogenous miRNAs were mainly presented in B
cells [25]. Last,
d. miRNA induced silencing complex (miRISC)-related
pathway that can interact with assembly proteins to form a
complex called miRISC [26] (Figure 1).
nSMase2, Neural Sphingomyelinase 2; hnRNPAs, Heterogeneous Nuclear Ribonucleoproteins; Ago2, Argonaute protein-2; ESCRT, endosomal sorting complexes required for transport; MBV, multivesicular body
a. nSMase2 increase the number of exosomal miRNAs
b. hnRNPA-1, a specific heterogeneous Nuclear
Ribonucleoprotein, bindings short sequence motif presents in
miRNAs 5’-3’ ends to guide their loading into exosomes
c. The 3’ uridylated ends of miRNAs are a “tag” for exosome
encapsulation
d. miRNA can interact with the Ago2 assembly, to form a
complex called miRISC for its internalization in the exosome
e. ESCRT machinery sequentially assembles on endosomes
where it generates MVB vesicles
Some profiling studies showed that the incorporation of miRNAs into exosome, is not a random process; in fact, several studies demonstrated the different miRNAs expression level in a variety of cell lines with those from their respective derived exosomes [27]. Similarly, Ohshima compared the expression levels of let-7 miRNA family members in exosomes derived from the gastric cancer cell line AZ-P7a with those from other cancer cell lines, including the lung cancer cell line SBC-3/DMS-35/NCI-H69, the colorectal cancer cell line SW480/SW620, and the stomach cancer cell line AZ-521. As a result, they found that members of the let-7 miRNA family are abundant in exosomes derived from AZ-P7a, but are less abundant in exosomes derived from other cancer cells [28]. Moreover, some reports have shown that exosomal miRNA expression levels are altered under different physiological conditions. The serum level of exosome containing miR-21 was lower in healthy donors than patients with glioblastoma [29]. Levels of let-7f, miR-20b, and miR- 30e-3p were lower in vesicles from plasma of non-small-cell lung carcinoma patients than normal controls. Different levels of eight exosomal miRNAs, including miR-21 and miR141, were also found between benign tumors and ovarian cancers [30].
between benign tumors and ovarian cancers [30]. According to these previous studies, a class of miRNAs are preferentially sorted into exosomes, such as members of the miR- 320 family that are widely distributed in exosomes derived from normal tissue and tumors [31]. In addition, some miRNAs, miR-451 for example, are highly expressed in exosomes derived from normal cells, such as the HMC-1 cell line and the HEK293T cell line [32]. In summary, miRNAs present specific sequences that may guide their incorporation into exosomes, whereas some enzymes or other proteins may control exosomal miRNAs sorting as well, in a miRNA sequence-independent fashion. The evidences highlight the role of miRNAs for the diagnosis and staging of BCa that could provide great improvements in clinical decision-making.
Discussion
MiRNAs in BCa
The complex regulatory mechanisms of miRNAs that cause the degradation of the target mRNA or the inhibition of translation, define the role of miRNAs as oncogenes or tumor suppressor genes. In this contest, downregulated miRNAs that target tumor suppressor genes are considered to be onco-miRNAs, and upregulated miRNAs that target oncogenes are considered to be tumor-suppressing miRNAs [33]. Similar to other malignancies, BCa is characterized by an alteration of the molecular profile caused by the accumulation of genetic and epigenetic modification. Several reports compared the presence of circulating miRNAs resulted from bladder cells undergoing necrosis and apoptosis with modifications of the normal bladder phenotype [34]. For these reasons, miRNAs related to bladder carcinogenesis were gradually explored, raising the interest of several research groups in using miRNA as tools for diagnosis, prediction of tumor progression and BCa metastasis [35]. A recent work performed a global miRNA profiling of 392 serum samples of BCa patients with 100 non-cancer samples and 480 samples of other types of cancer as controls by a miRNA microarray [36]. They identified 7 miRNA candidates (miR-6087, miR-6724-5p, miR-3960, miR-1343-5p, miR-1185-1- 3p, miR-6831-5p and miR-4695-5p) that clearly discriminated BCa from non-cancer controls and other types of cancer, regardless of cancer stage and grade, enabling the specific and early detection of BCa with high accuracy. Among these, miR-6087 showed high diagnostic performance for BCa screening [36]. MiR-141 and miR- 205 are poor prognostic biomarkers of overall survival in BCa [37], whereas severely decrease of miR-29c∗ is able to discriminated 50% of NMIBC in advanced cancer [38]. The expression profiles of miRNAs by either microarray or deep sequencing technologies, have been documented in numerous investigations confirming their putative role as tumor marker for BCa diagnosis [39,40].
As example, miR-145, miR-143, and miR-125b are downregulated in BCa tissues and are defined tumor-suppressing miRNAs, whereas miR-183, miR-96, miR-17-5p, and miR-20a are oncogenic miRNAs because of their upregulation in BCa tissues [41]. Recent research showed that miRNA-mediated post-transcriptional regulation plays an important role in chemoresistance and can affect drug efficacy by regulating the expression of multiple drugresistance- related proteins [42]. The 5637 and H-bc cell lines are the most multichemosensitive and resistant BCa cell lines, respectively. Lv and coworkers demonstrated that 82 miRNAs were differently expressed between 5637 cells and H-bc cells [43]. Moreover, a microarray analysis of gemcitabine-resistant revealed 66 differentially expressed miRNAs in BCA cell lines; among these, miR-1290 and miR-138 showed increased expression levels in gemcitabine-resistant cells, while let-7b and let-7i exhibited decreased expression [44]. Concerning the drug-resistance-related miRNAs, Adam and colleagues showed that the stable expression of miR-200 in mesenchymal UMUC3 cells increased E-cadherin levels and sensitivity to EGFR blockers (cetuximab) and decreased the expression of zinc finger E-Box binding Homeobox (ZEB) 1, 2, as well as cell migration [45]. Since then, studies have focused on the role of miRNAs in the chemoresistance of BCa. For example, increased expression level of miR-203 enhanced the sensitivity of BCa cells to cisplatin by promoting cell apoptosis [46]. Based on these preclinical studies, clinical research on miRNA-targeted therapeutic strategies will open a new chapter in BCa cancer therapy.
MiRNAs in Several Body Fluids Related to BCa
Advanced technology uses liquid biopsy for the analyses of circulating tumor cells (CTCs), exosomes, and circulating miRNAs in patients’ blood and urine as samples instead of primary BCa tissue [47]. Several cell-free miRNAs, such as miR-497 and miR-214 were detected by liquid biopsy in plasma and urine of BCa patients, respectively [48,49]. Although this application could be promising for the discovery of novel circulating biomarkers, liquid biopsy in BCa treatment is yet to be explored and, at the moment, is used only for diagnosing BCa.
Recent studies demonstrated that miRNAs are abundantly present in body fluid including serum, plasma, cerebrospinal fluid, saliva and urine and their expression is correlated with several diseases, including cancer [50]. Cells undergoing necrosis and apoptosis are the major source of circulating miRNAs. Tumor cells actively secrete cellular elements that can be detected in body fluids as either free circulating miRNAs or bound to ribonucleoprotein complexes and encapsulated in extracellular vesicles such as exosomes [51]. MiRNAs in exosomes may support tumor angiogenesis, invasion and metastasis through bone marrow-derived cells mobilization [52]. This encapsulated transport, complexed with proteins or lipids, protects miRNAs from degradation and is stable in body fluids. Thank to miRNAs capacity to be easily screened and precisely quantified by a variety of standard methods like qRT-PCR, in situ hybridization, enzymatic luminescence miRNA assay, microarray or next-generation sequencing [53], they can be clinically applied to detect tumors at an early stage. The stability, the frequency, the reversibility and the accessibility in body fluids of miRNAs as well as the resistance to various storage conditions and stability up to 24 h storage, make miRNAs optimal biomarker candidates of neoplastic diseases [54,55]. Even though total circulating miRNA levels are increased in cancer patients [56], the fraction of cancer-specific miRNAs is low and needs to be quantified compare to the background of “normal” miRNAs, thereby pointing out the need for sensitive and specific detection methods.
A wide range of experimental data demonstrated the oncogenic role of miRNAs in the BCa, through influencing tumor growth, cell survival and angiogenesis.
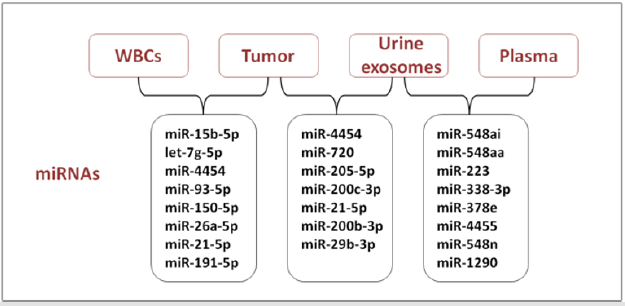
Evaluation of miRNA pattern expression as a potential tool for the early diagnosis and prognosis of BCa is a challenge in urooncologic research, and it could provide benefits for invasiveness and cost reduction in health care services [57]. Recently, a greater amount of data on this subject were collected and most studies use urine samples to investigate diagnostic or prognostic miRNA markers in BCa. Amstrong and colleagues analyzed miRNAs of matched formalin-fixed paraffin-embedded (FFPE)-tumor tissue, plasma, urine exosomes, white blood cells fraction (WBCs) from 16 patients with NMIBC, using Nanostring miRNA assays [58]. They identified 25 miRNAs in each of the bio-specimen sources, presenting an overlap between and amongst the different biological matrices (Figure 2).
MiR-4454 and miR-21-5p were both detectable in FFPE-tumor
tissue, urine exosomes and WBCs, while let-7a-5p, let-7b-5p, miR-
451a and miR-16 were abundant in FFPE-tumor tissue, WBCs and
plasma [58]. Moreover, an evaluation of the useful non-invasive
biomarkers of low-grade tumors (Ta), was examined in matched
bio-specimens from Ta patients only, between FFPE-tumor tissue
and urine exosomes. The highest abundance of miR-4454, miR-
720/3007a and miR-21 were found in both bio-specimen sources.
Zhou and coworkers, identified miR-4454 gene as target of
nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB)
transcriptional activity in TNFα-stimulated HeLa Cells, by using
ChIP-Seq, Genechip and miRNA-Seq techniques [59]. Remarkably,
the stimulation of endogenous NF-κB activity by TNFα and
siRNA interference of NF-κB p65, the protein inhibitor of NF-κB
transcriptional activity, resulted in a significant increase of miR-
4454 [59]. Recent literature clarified the role of miR-720/3007a
as tRNA-derived fragments (tRFs), firstly identified in NMIBCs
[60]. tRFs are 14-32 base long single-stranded RNA derived from
mature or precursor tRNA. They are grouped into 3 classes (tRF-
1, -3, and -5) and tRFs can be divided into 5 tiRNA (tRNA halves)
and 3 tiRNA depending on the sources. They play important roles
in tumorigenesis, promoting both cell proliferation and cell cycle
progression by regulating the expression of oncogenes. Guzman
et al showed the correlation between cellular expression levels of
five miRNAs (miR-21, let-7a, miR-100, and miR-125b and miR-205)
and two tRNA-derived miRNAs, miR-720 and miR-1274b, in MCF7
cells, compared with those in extracellular vesicle (EVs) levels [61].
Interestingly, while correlations in the cellular expression of miR 125b, miR-100, and let-7a are mirrored in the EVs, tRNA-derived
miRNA levels are found only in the EVs. These findings suggest the
possible use of tRNA-derived small RNA fragments in combination
with known miRNA signatures of tumors to distinguish circulating
tumor-derived EVs from physiological EVs derived from other cell
sources.
An important miRNA profiling investigation was conducted
on 23-paired samples, normal and tumoral bladder tissue, by
Braicu and colleagues [62]. They identified 18 downregulated
and 187 upregulated miRNAs, by next-generation sequencing
investigation, focalizing on a systematic identification of altered
miRNAs and genes mutated involved in BCa tumorigenesis and
progression. Among all these miRNAs, relevant findings were
obtained concerning miR-21. As a miRNA commonly upregulated
in human cancers, in vitro models demonstrated that miR-21
is able to regulate cell proliferation and migration via its cross
talk with phosphatase and tensin homolog (PTEN) and tumor
protein 53 (TP53), one of the most important pathways related to
bladder carcinogenesis [63]. Moreover, a recent study confirmed
the altered expression level for miR-21, miR-205 and miR-200c
in BCa analyzing data generated by pairing the tissue to plasma
samples [64]. Studies on chondrogenic differentiation of murine
mesenchymal stem cells (MSCs) demonstrated that miR-145 is a
negative regulator through directly targeting SRY-Box transcription
factor 9 (SOX9), a key transcription factor for chondrogenesis
[65]. Remarkably, SOX9 was significantly upregulated in tissues of
patients with either renal cell carcinoma or BCa and correlated to
the advanced pathological grade and clinical stage of patients [66].
Although there are no studies that confirm the correlation between
miR-145 and proteins involved in cell differentiation such as SOX9
in BCa, Hashimoto and coworkers confirmed that miR-145 and
miR-133 directly control fascin actin-bundling protein 1 (FSCN1),
an actin-binding protein required for the formation of actin-based
cell-surface protrusions and cytoplasmic bundles of microfilaments
in BCa patients [67]. In summary, miR-145 and miR-133a might
function as tumor suppressors through direct repression of FSCN1
in BCa.
The need to discriminate patients with BCa from those with
benign diseases such as glomerulonephritis, benign prostate
hyperplasia, infection, or urinary calculi lead Piao and colleagues to
perform a multicenter study using 543 urine samples obtained from
the National Biobank of Korea, including 326 BCa, 174 hematuria
and 43 pyuria without cancer [68]. In this study, the expression
ratio between urinary cell-free miRNAs, miR-6124 and miR-4511,
was found to be significantly higher in patients with BCa than in
those with hematuria and pyuria. Although these miRNAs ranged
a sensitivity and specificity of 78.5% and 74.1%, respectively,
were not sensitive enough to replace cystoscopy; in fact, avoiding
unnecessary cystoscopies by an accurate, cost-effective and simple
urinary test remains an unresolved issue. Although the role of
miRNAs in oncologic pathways is under evaluation, the research of
other promising miRNA candidates in BCa, may open the way to
novel approaches for diagnosis and prognosis.
Conclusion
Recent studies have identified the role played by epigenetic mechanisms, such as DNA methylation, histone modifications and miRNA inhibitory activity during bladder carcinogenesis. The dysregulation of these mechanisms may increase the susceptibility to pathological conditions, suggesting a potential role of epigenetic factors as pharmacological targets to restore the homeostatic regulation of cancer diseases. Considering the real need to develop new, simple and accurate molecular markers for the early diagnosis and prognosis of BCa, miRNAs could have valuable importance as pathological predictors in biological fluids, as well as in tissues. For these reasons, clinical studies confirming the pivotal role of epigenetic modulators in BCa could be decisive for the diagnosis and treatment of diseases in clinical activity.
Acknowledgment
Fondazione CARIT.
Conclusion
The authors declare no conflict of interest.
References
- Park JC, Citrin DE, Agarwal PK, Apolo AB (2014) Multimodal management of muscle-invasive bladder cancer. Curr Probl Cancer 38(3): 80-108.
- Têtu B (2009) Diagnosis of urothelial carcinoma from urine. Mod Pathol 22(2): S53-S59.
- Van Rhijn BWG, van der Poel HG, van der Kwast TH (2005) Urine markers for bladder cancer surveillance: A systematic review. Eur Urol 47(6): 736-748.
- Zhang S, Wang Y, Bondaruk J, Majewski T, Yao H, et al. (2019) Detection of Bladder Cancer in Urine Sediments by a Novel Multicolor Fluorescence In Situ Hybridization (Quartet) Test. Eur Urol Focus 5(4): 664-675.
- Zhou L, Yang K, Li X, Ding Y, Mu D, et al. (2019) Application of fluorescence in situ hybridization in the detection of bladder transitional-cell carcinoma: A multi-center clinical study based on Chinese population. Asian J Urol 6(1): 114-121.
- Marin‐Aguilera M, Mengual L, Ribal MJ, Burset M, Arce Y, et al. (2007) Utility of a multiprobe fluorescence in situ hybridization assay in the detection of superficial urothelial bladder cancer. Cancer Genet Cytogenet 173(2): 131-135.
- Cha EK, Tirsar LA, Schwentner C, Christos PJ, Mian C, et al. (2012) Immunocytology is a strong predictor of bladder cancer presence in patients with painless hematuria: a multicentre study. Eur Urol 61(1): 185-192.
- Eissa S, Swellam M, Ali‐Labib R, Mansour A, El‐Malt O, et al. (2007) Detection of telomerase in urine by 3 methods: Evaluation of diagnostic accuracy for bladder cancer. J Urol 178(3 Pt 1): 1068-1072.
- Glas AS, Roos D, Deutekom M, Zwinderman AH, Bossuyt PM, et al. (2003) Tumor markers in the diagnosis of primary bladder cancer. A systematic review. J Urol 169(6): 1975-1982.
- Moll R, Lowe A, Laufer J, Franke WW (1992) Cytokeratin 20 in human carcinomas. A new histodiagnostic marker detected by monoclonal antibodies. Am J Phatol 140(2): 427-447.
- Guo B, Luo C, Xun C, Xie J, Wu X, et al. (2009) Quantitative detection of cytokeratin 20 mRNA in urine samples as diagnostic tools for bladder cancer by real-time PCR. Exp Oncol 31(1): 43-47.
- Yildiz IZ, Recavarren R, Armah HB, Bastacky S, Dhir R, et al. (2009) Utility of a dual immunostain cocktail comprising of p53 and CK20 to aid in the diagnosis of non-neoplastic and neoplastic bladder biopsies. Diagn Pathol 4: 35.
- Mumtaz S, Hashmi AA, Hasan SH, Edhi MM, Khan M (2014) Diagnostic utility of p53 and CK20 immunohistochemical expression grading urothelial malignancies. Int Arch Med 7(1): 36.
- Song MS, Rossi JJ (2017) Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem J 474(10): 1603-1618.
- Ling H, Fabbri M, Calin GA (2013) MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 12(11): 847-865.
- Esquela-Kerscher A, Slack FJ (2006) Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6(4): 259-269.
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043): 834-838.
- Bose M, Bhattacharyya SN (2016) Target-dependent biogenesis of cognate microRNAs in human cells. Nat Commun 7: 12200.
- Fare M, Yeom KH, Haagsma AC, Chauhan S, Heo I, et al. (2016) TRBP ensures efficient Dicer processing of precursor microRNA in RNA-crowded environments. Nat Commun 7: 13694.
- Iorio MV, Croce CM (2012) MicroRNA involvement in human cancer. Carcinogenesis 33(6): 1126-1133.
- Henne WM, Buchkovich NJ, Emr SD (2011) The ESCRT pathway. Dev Cell 21(1): 77-91.
- Zhang J, Li S, Li L, Li M, Guo C, et al. (2015) Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 13(1): 17-24.
- Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, et al. (2013) Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem 288(15): 10849-10859.
- Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, et al. (2013) Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4(1): 2980.
- Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, et al. (2014) Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep 8(6): 1649-1658.
- Frank F, Sonenberg N, Nagar B (2010) Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465(7299): 818-822.
- Guduric-Fuchs J, O’Connor A, Camp B, O’Neill CL, Medina RJ, et al. (2012) Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics 13: 357.
- Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, et al. (2010) Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One 5(10): e13247.
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, et al. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10(12): 1470-1476.
- Schwarzenbach H, Gahan PB (2019) MicroRNA Shuttle from Cell-To-Cell by Exosomes and Its Impact in Cancer. Noncoding RNA 5(1): 28.
- Whiteside TH (2016) Exosomes and tumor-mediated immune suppression. J Clin Invest 126(4): 1216-1223.
- Abak A, Abhari A, Rahimzadeh S (2018) Exosomes in cancer: small vesicular transporters for cancer progression and metastasis, biomarkers in cancer therapeutics. Peer J 6: e4763.
- Cai Z, Zhang F, Chen W, Zhang J, Li H (2019) MiRNAs: A Promising Target in the Chemoresistance of Bladder Cancer. Onco Targets Ther 12: 11805-11816.
- Ellinger J, Müller SC, Dietrich D (2015) Epigenetic biomarkers in the blood of patients with urological malignancies. Expert Rev Mol Diagn 15(4): 505-516.
- Truta A, Popon TAH, Saraci G, Ghervan L, Pop IV (2016) Novel non invasive diagnostic strategies in bladder cancer. Clujul Med 89(2): 187-192.
- Usuba W, Urabe F, Yamamoto Y, Matsuzaki J, Sasaki H, et al. (2019) Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci 110(1): 408-419.
- Braicu C, Cojocneanu-Petric R, Chira S, Truta A, Floares A, et al. (2015) Clinical and pathological implications of miRNA in bladder cancer. Int J Nanomedicine 10: 791-800.
- Rosenberg E, Baniel J, Spector Y, Faerman A, Meiri E, et al. (2013) Predicting progression of bladder urothelial carcinoma using microRNA expression. BJU Int 112(7): 1027-1034.
- Han Y, Chen J, Zhao X, Liang C, Wang Y, et al. (2011) MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One 6(3): e18286.
- Tolle A, Jung M, Rabenhorst S, Kilic E, Jung K, et al. (2013) Identification of microRNAs in blood and urine as tumour markers for the detection of urinary bladder cancer. Oncol Rep 30(4): 1949-1956.
- Nagata M, Muto S, Horie S (2016) Molecular Biomarkers in Bladder Cancer: Novel Potential Indicators of Prognosis and Treatment Outcomes. Dis Markers 2016: 8205836.
- Boguslawska J, Kryst P, Poletajew S, Piekielko-Witkowska A (2019) TGF-β and microRNA Interplay in Genitourinary Cancers. Cells 8(12): 1619.
- Lv L, Deng H, Li Y, Zhang C, Liu X, et al. (2014) The DNA methylation-regulated miR-193a-3p dictates the multi-chemoresistance of bladder cancer via repression of SRSF2/PLAU/HIC2 expression. Cell Death Dis 5(9): e1402.
- Kozinn SI, Harty NJ, Delong JM, Deliyiannis C, Logvinenko T, et al. (2013) MicroRNA Profile to Predict Gemcitabine Resistance in Bladder Carcinoma Cell Lines. Genes Cancer 4(1-2): 61-69.
- Adam L, Zhong M, Choi W, Qi W, Nicoloso M, et al. (2009) MiR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res 15(16): 5060-5072.
- Li P, Yang X, Cheng Y, Zhang X, Yang C, et al. (2017) MicroRNA-218 Increases the Sensitivity of Bladder Cancer to Cisplatin by Targeting Glut1. Cell Physiol Biochem 41(3): 921-932.
- Busetto GM, Ferro M, Del Giudice F, Antonini G, Chung BI, et al. (2017) The Prognostic Role of Circulating Tumor Cells (CTC) in High-risk Non–muscle-invasive Bladder Cancer. Clinical Genitourin Cancer 15(4): e661-e666.
- Du M, Shi D, Yuan L, Li P, Chu H, et al. (2015) Circulating miR-497 and miR-663b in plasma are potential novel biomarkers for bladder cancer. Sci Rep 5: 10437.
- Wang J, Zhang X, Wang L, Dong Z, Du L, et al. (2015) Downregulation of urinary cell-free microRNA-214 as a diagnostic and prognostic biomarker in bladder cancer. J Surg Oncol 111(8): 992-999.
- Foye C, Yan IK, David W, Shukla N, Habboush Y, et al. (2017) Comparison of miRNA quantitation by Nanostring in serum and plasma samples. PLoS One 12(12): e0189165.
- Tölle A, Blobel CC, Jung K (2018) Circulating miRNAs in blood and urine as diagnostic and prognostic biomarkers for bladder cancer: an update in 2017. Biomark Med 12(6): 667-676.
- Wang Z, Chen JQ, Liu JL, Tian L (2016) Exosomes in tumor microenvironment: novel transporters and biomarkers. J Transl Med 14: 297.
- Dave VP, Ngo TA, Pernestig AK, Tilevik D, Kant K, et al. (2019) MicroRNA amplification and detection technologies: opportunities and challenges for point of care diagnostics. Lab Invest 99(4): 452-469.
- Liu X, Liu X, Wu Y, Wu Q, Wang Q, et al. (2017) MicroRNAs in biofluids are novel tools for bladder cancer screening. Oncotarget 8(19): 32370-32379.
- Takahashi RU, Prieto-Vila M, Kohama I, Ochiya T (2018) MicroRNA in Body Fluids - Development of the Novel Plat Form for Cancer Therapeutics and Diagnosis. Gan to Kagaku ryoho 45(6): 899-905.
- Cui M, Wang H, Yao X, Zhang D, Xie Y, et al. (2019) Circulating MicroRNAs in Cancer: Potential and Challenge. Front Genet 10: 626.
- Poli G, Brancorsini S, Cochetti G, Barillaro F, Egidi MG, et al. (2015) Expression of inflammasome-related genes in bladder cancer and their association with cytokeratin 20 messenger RNA. Urol Oncol 33(12): 505.e1-505.e7.
- Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ (2015) MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer 14: 194.
- Zhou F, Wang W, Xing Y, Wang T, Xu X, et al. (2014) NF-kappaB target microRNAs and their target genes in TNFalpha-stimulated HeLa cells. Biochim Biophys Acta 1839(4): 344-354.
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, et al. (2010) Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16(4): 673-695.
- Guzman N, Agarwal K, Asthagiri D, Yu L, Saji M, et al. (2015) Breast Cancer-Specific miR Signature Unique to Extracellular Vesicles includes “microRNA-like” tRNA Fragments. Mol Cancer Res 13(5): 891-901.
- Braicu C, Buiga R, Cojocneanu R, Buse M, Raduly L, et al. (2019) Connecting the dots between different networks: miRNAs associated with bladder cancer risk and progression. J Exp Clin Cancer Res 38(1): 433.
- Calderaro J, Rebouissou S, De Koning L, Masmoudi A, Herault A, et al. (2014) PI3K/AKT pathway activation in bladder carcinogenesis. Int J Cancer 134(8): 1776-1784.
- Dong F, Xu T, Shen Y, Zhong S, Chen S, et al. (2017) Dysregulation of miRNAs in bladder cancer: altered expression with aberrant biogenesis procedure. Oncotarget 8(16): 27547-27568.
- Yang B, Guo H, Zhang Y, Chen L, Ying D, et al. (2011) MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting SOX9. PLoS One 6(7): e21679.
- Wan YP, Xi M, He HC, Wan S, Hua W, et al. (2017) Expression and Clinical Significance of SOX9 in Renal Cell Carcinoma, Bladder Cancer and Penile Cancer. Oncol Res Treat 40(1-2): 15-20.
- Hashimoto Y, Skacel M, Adams JC (2005) Roles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker?. Int J Biochem Cell Biol 37(9): 1787-1804.
- Piao XM, Jeong P, Kim YH, Byun YJ, Xu Y, et al. (2019) Urinary cell‐free microRNA biomarker could discriminate bladder cancer from benign hematuria. Int J Cancer 144(2): 380-388.

 Mini Review
Mini Review