Abstract
DHs, meaning that the chromosomes doubled haploids, are defined as 100% homozygous. In this study, three DH lines and one highly inbred line were used to analysis the heterozygosity ratio, and interesting finding that the centromere sequences of doubled haploids were heterozygous. Nevertheless, it was difficult to explain why the centromere sequences were heterozygous in DH lines? This finding would provide new insight for basic and applied research scientists
Keywords: Brassica rapa; Doubled Haploid; Centromere; Heterozygosity
Introduction
Hybrid vigor, or heterosis, means that the first-generation hybrids show the better performance than each parent, which is a biological phenomenon among crop production. In the last few decades, hybrid vigor has been widely reported in many crops, including lots of the field crops (Oryza sativa, Zea mays, and so on) and a series of vegetables (Brassica rapa, Cucumis sativus, and so on) Song et al. [1-3], and there are also animals, including Canis lupus familiaris (dogs) Proops et al. [4,5] And the highly inbred lines and doubled haploid (DH) played a key role for heterosis, especially, in basic and applied research, as well as to apply in breeding programs Germana et al. [6,7] In most flowering plants, the immature microspores could be used to develop into embryos and, subsequently, into plants in vitro for DH lines Ferrie et al. [8] DH lines, meaning that the chromosomes doubled haploids (2n), are defined as 100% homozygous. However, maybe it was not 100% homozygous. In our study, we found that the centromere sequences of the DH lines in Brassica rapa were heterozygous.
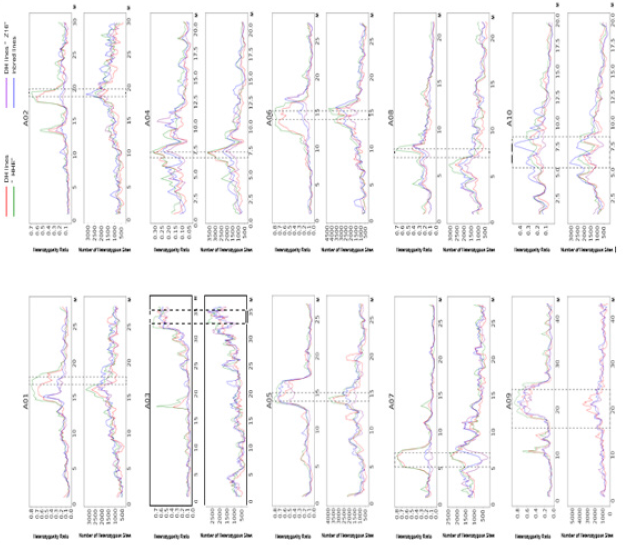
Three DH lines (HHE, XJD4, and Z16) and one highly inbred line (YTBJ) were used to re-sequence (30x) and detect variants compared with the Brassica rapa reference genomes version 3.0. The Genome Analysis TK (GATK 0.2.4.3) software was used to call variants, and the genotypes of every position were analyzed. ‘0’ represents the reference allele, and ‘1’ represents the variant allele. ‘0/0’ represents that this position was homozygous, which consistent with the reference genome, and ‘0/1’ represents that this position was heterozygous, which contains two genotypes (reference genome type and the first variant type). ‘1/1’ represents that this position was homozygous, which consistent with the variant type. In this study, the number of heterozygous sites (‘0/1’ genotypes) and heterozygosity ratio were counted (Figure 1) from the chromosomes A01 to A10. Sliding window statistical method was used to analyze the variants. The sliding window size was 100 kb, and the step size was 1 Mb. Heterozygosity ratio = (the numbers of heterozygous sites in the sliding window / the total number of variants) * 100%.
Figure 1: Analysis of the heterozygosity ratio and the number of heterozygous sites from chromosomes A01 to A10 in Brassica rapa.
The physical positions of centromere refered to the Brassica rapa reference genome 3.0 Zhang [9]. The three DH lines (HHE, XJD4, and Z16) and one highly inbred line (YTBJ) were genetically stable lines for generating hybrid, and the phenotypic traits were no longer isolated. However, the centromere sequences were heterozygous from 20% to 80% in A01 to A10 in the HHE, XJD4, Z16, and YTBJ lines (Figure 1), and the imaginary line regions were the centromere regions in Fig. 1. The numbers of heterozygous sites from the centromere region were significantly higher than other regions in the chromosomes (Figure 1). In A01/05/06/07/09, the heterozygosity of the centromere sequences was higher up to 80%, and there was only one the heterozygous peak in the chromosomes A05/06/07. However, in the A02/04/08/09, there were more than one heterozygous peak, and besides the centromere regions, there was one more heterozygous peak in the non-centromere regions. From Figure 1, the tendency of the number of heterozygous sites and heterozygosity ratio were almost same, except for the centromere regions of the chromosomes A02/06, which implied that the total number of sequencing sites at different location was various.
But it could not explain why the centromere sequences were heterozygous in DH lines. Why were the centromere sequences of the DH lines heterozygous? So far, there was no reliable evidence. There may be several possibilities. First, the centromere sequences of the DHs were not assembled well. With the development of the next-generation sequencing, lots of crop genomes were assembled Metzker [10]. However, some genome regions still remain recalcitrant and were not very well known. For example, there are lots of repeat sequences, and it was difficult to assemble the large repetitive regions under the existing technologies Metzker [10]. And the large of repetitive sequences could hide lots of key information, like centromere. Notoriously, it was very difficult to assemble the centromere sequences, and the high density of repetitive sequences (transposon elements) in these regions makes them assembly problematic Rudd et al. [11]. Second, there are so many transposon elements (TEs) and retro-transposon elements (Retro-TEs) in the centromere regions, and the transposons keep bouncing, which would result in the increasing of the heterozygosity. These long terminal repeat retrotransposons (LTR-RTs) burst events were not inherited from a common ancestor, but instead were speciesspecific bursts that occurred after the divergence of Brassica species Cai et al. [12].
Third, because of a large number of repetitive sequences, when mapping genes in the centromere regions, it was difficult to fine map the genes location. So, this finding would provide new insight for basic and applied research scientists.
Ethics Approval and Consent to Participate
Not Applicable
Consent for Publication
Not Applicable
Availability of Data and Materials
Not Applicable
Competing Interests
The authors declare that they have no competing interests.
Funding
This work was funded by the National Key Research and Development Program of China (2016YFD0101701) and the National Natural Science Foundation of China (31772302).
Authors’ Contributions
RS, FL, LY, and GL found the phenomenon. GL and LY drafted the manuscript. SZ, HZ, and SZ helped to analysis the phenomenon. All the authors approved the manuscript.
Acknowledgment
Thanks to the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, Beijing, China.
References
- Song G, Zhai H, Peng Y, Zhang L, Wei G, et al. (2010) Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Molecular plant 3(6): 1012-1025.
- Ko D, Rohozinski D, Song Q, Taylor S, Juenger T, et al. (2016) Temporal shift of circadian-mediated gene expression and carbon fixation contributes to biomass heterosis in maize hybrids. PLoS genetics 12(7): e1006197.
- Saeki N, Kawanabe T, Ying H, Shimizu M, Kojima M, et al. (2016) Molecular and cellular characteristics of hybrid vigour in a commercial hybrid of Chinese cabbage. BMC plant biology 16(1): 45.
- Proops L, Burden F, Osthaus B (2009) Mule cognition: a case of hybrid vigour? Animal cognition 12(1): 75-84.
- Nicholas F, Arnott E, Mc Greevy P (2016) Hybrid vigour in dogs? The Veterinary Journal 214: 77-83.
- Germana M (2011) Gametic embryogenesis and haploid technology as valuable support to plant breeding. Plant cell reports 30(5): 839-857.
- Dwivedi S, Britt A, Tripathi L, Sharma S, Upadhyaya H, et al. (2015) Haploids: constraints and opportunities in plant breeding. Biotechnology advances 33(6): 812-829.
- Ferrie A, Caswell K (2011) Isolated microspore culture techniques and recent progress for haploid and doubled haploid plant production. Plant Cell, Tissue and Organ Culture 104(3): 301-309.
- Zhang Lei (2018) Improved reference genome and chromatin interactions analysis in Brassica rapa. PhD dissertation. China Agricultural University, Beijing.
- Metzker M (2010) Sequencing technologies-the next generation. Nature reviews genetics 11(1): 31-46.
- Rudd M, Willard H (2004) Analysis of the centromeric regions of the human genome assembly. Trends in genetics 20(11): 529-533.
- Cai X, Cui Y, Zhang L, Wu J, Liang J, et al. (2018) Hotspots of Independent and Multiple Rounds of LTR-retrotransposon Bursts in Brassica Species. Horticultural Plant Journal 4(4): 165-174.

 Short Communication
Short Communication