Abstract
Riboswitches are mostly cis-acting gene control elements residing at the 5-prime untranslated region of mRNAs in bacteria and archaea. Riboswitches control gene expression by directly binding their ligands without the need for any protein factors. Among the nearly 40 distinct riboswitch classes discovered, for the time being, there is only one riboswitch class with the dual function of both a self-cleaving ribozyme and a metabolite-responsive riboswitch. The glucosamine-6-phosphate-sensing ribozyme cleaves its mRNA upon binding to glucosamine-6-phosphate, which serves as a co-factor. As a result, it controls the gene expression of glmS mRNA as a riboswitch. Therefore, the translation of the enzyme glucosamine-6-phosphate synthase, essential for the synthesis of glucosamine-6-phosphate, is inhibited when there is enough glucosamine-6-phosphate in the bacterial cell. Glucosamine-6-phosphate is a key precursor for cell-wall synthesis mainly in Gram-positive bacteria. Therefore, glucosamine-6-phosphate’s analogs that trigger self-cleavage of the glmS ribozyme may be used for antibacterial drug discovery. Here, we present a mini-review of currently known glmS riboswitch activators, which can play a role in substituting glucosamine-6-phosphate and possibly inhibiting pathogenic bacteria.
Keywords: glmS; Riboswitch; Ribozyme; Activators; Substitute; Pathogenic Bacteria; Drug Discovery
Abbreviations: RNA: Ribonucleic Acid; GlcN6P: Glucosamine-6-Phosphate; HTS: High-Throughput Assays; GlcN- Glucosamine; Glc6P- Glucose-6-Phosphate; Glc- Glucose; CGlcN: carba-α-D-glucosamine; CGlcN6P: Carba-α-D-glucosamine-6-phosphate
Introduction
Riboswitches are regulatory RNA elements, which are usually found in the 5’-untranslated regions (UTR) of the gene in many bacteria. They regulate gene expression by directly binding a specific ligand [1]. Most of the riboswitches are found using a combination of bioinformatics approaches and experimental methods such as inline probing of RNAs. The bioinformatics approaches including a search for conserved sequences (motifs) in non-coding regions of RNAs and their alignment with specialized software [2]. As a result of the bioinformatics research, riboswitch distribution throughout the three domains is almost exclusively found in the domain Bacteria and Archaea and is very rare in Eukaryota [3]. Bacterial riboswitches are potential targets for antibacterial drug discovery, and as such the glmS riboswitch presents a prospective target for novel antibacterial compounds [4,5].
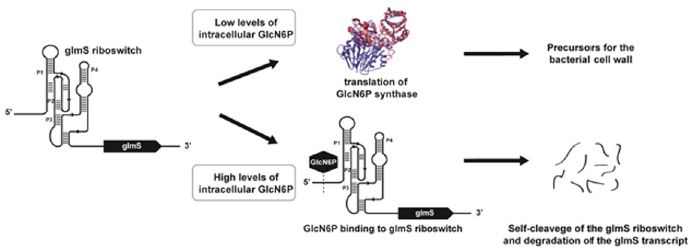
The glmS riboswitch, also known as a glucosamine-6-phosphate activated ribozyme, is the first discovered naturally occurring catalytic RNA, which can sense a small molecule as a coenzyme [6]. It controls gene expression by mRNA destabilization by selfcleavage. This unique characteristic makes the glmS ribozyme also a riboswitch the only discovered dual-functioning riboswitch/ ribozyme class. The specific ligand that is being recognized of the glmS riboswitch is glucosamine-6-phosphate (GlcN6P), which in cases of absence or a very low concentration does not activate the glmS riboswitch and the glmS mRNA is successfully translated into the enzyme glucosamine 6-phosphate synthase (Figure 1). The latter catalyzes the synthesis of the GlcN6P, which is an essential precursor for the cell wall in bacteria. When the concentration of GlcN6P is high enough for the bacteria on a micromoles scale, the glmS ribozyme cleaves itself and down-regulates the glmS gene expression. Hence, GlcN6P synthase is not translated.
Figure 1: Mechanism of the glmS riboswitch regulation. At low levels or full absence of intracellular glucosamine-6-phosphate (GlcN6P) in the bacterial cell, the enzyme glucosamine-6-phosphate synthase is translated and used for catalytic regulation in the biochemical pathway for synthesis of peptidoglycan. GlcN6P acts as a co-factor of the glmS ribozyme, which as a result goes through self-cleavage and leads to degradation of the glmS mRNA.
The secondary structure оf the glmS riboswitch consists of 4 highly conservative paired regions (P1-P4). The mechanism by which the glmS riboswitch is self-cleaved is suggested to be due to the GlcN6P binding to a pre-folded active site pocket (ligandbinding pocket) and direct catalysis of the process with the support of Mg2+ [7]. Another research by Jansen et al. also suggests the direct involvement of the ligand as the cleavage site is at the 5′-end of the catalytic core of the riboswitch [8]. The ribozyme catalyzes transesterification, which is dependent upon the availability of the amine group in GlcN6P and its acid dissociation constant. As a result, there are 5′-hydroxyl and 2′,3′-cyclic phosphate termini of the cleavage products which do not integrate the ligand [9]. The free 5’- hydroxyl group is recognized by the bacterial RNase J and the latter degrades the glmS mRNA.
Current Glms Riboswitch Activators
As already mentioned, glmS ribozyme/riboswitch is programmed to sense the presence of the enzyme GlcN6P and selectively bind to it which leads to its self-cleavage and degradation of the glmS mRNA (Figure 1). This mechanism of gene regulation can be used for the development of novel anti-riboswitch drugs by substituting the original glmS cofactor/ligand. Some of the approaches in the research and development of new binding ligands include performing high-throughput assays (HTS), screening libraries with hundreds of small therapeutic chemicals [10,11]. Such assay was performed among 960 bioactive compounds, as only 5 were considered active [11]. To be considered as such, their fluorescence had to be more than 10 standard deviations higher in comparison with the mean of the fluorescence intensity in the control probes in the absence of the GlcN6P ligand. However, after retesting only 1 was found to successfully activate the riboswitch self-cleavage and that was the cofactor analog glucosamine. Glucosamine (GlcN) can activate the glmS riboswitch self-cleavage, while glucose-6-phosphate (Glc6P) and glucose (Glc) cannot, suggesting the partially dispensable phosphate moiety of GlcN6P and confirming the essential role of the anime group. However, glucosamine is a weaker activator in comparison with GlcN6P due to its inability to interact with the N1 of G1 through a phosphate. Meanwhile, it has been demonstrated that b-hydroxylamines such as TRIS can also activate self-cleavage in the bacteria B. cereus. This is possible due to the presence of amine and hydroxyl groups in a proper orientation [12].
The prospect of using glmS riboswitch activators as antibacterial agents has been proven by investigating alternative compounds, demonstrating the antibacterial activity of carba-α-D-glucosamine (CGlcN), and carba-α-D-glucosamine-6-phosphate (CGlcN6P) in the highly pathogenic bacteria Staphylococcus aureus [13]. Also, the same team discovered that fluoro-carba-sugars such as (5aR) ‐ Fluoro‐carba‐α‐D‐glucosamine‐6‐phosphate are also glycomimetic activators of the glmS riboswitch [14]. They mimic the natural sugar conformations and successfully activate the self‐cleavage mechanism in glmS in both Bacillus subtilis and Staphylococcus aureus. However, they are still less active than the non-fluorinated analogs in the research.
Discussion
Targeting various RNAs has emerged as one of the most promising approaches in the battle with antibiotics resistance in bacteria. For instance, riboswitches with their highly conserved sequences and often-essential roles in bacterial metabolism are considered as potential new targets. Performing high-throughput screening assays for drug-like compounds, including for glmS ribozyme activators, cannot always be a useful method as the currently available chemical libraries are of limited diversity. Even so, it is a good starting point for finding analogs, which activate the self-cleavage mechanism, and analyzing the reasons why some of them have higher activity than others. In addition to HTS, novel approaches might be more successful in the search for proantibacterial and highly active substitutes for the glmS ligand. Such techniques can be the application of the antisense technology of engineered phage therapy [15].
Conclusion
Some riboswitches, and particularly the glmS riboswitch, are being employed as new targets for the development of novel antibacterial agents. However, there are still not many discovered glmS substitute activators and hence inhibitors of the bacterial growth in vivo. Nevertheless, we strongly believe based on our current research that such inhibitors can be developed soon.
Acknowledgments
The research in Robert Penchovsky’s laboratory is also funded by a grant DN13/14/20.12.2017 awarded by the Bulgarian National Science Fund (BNSF).
Conflict of Interest
The authors decline any conflict of interest.
References
- Roth A, Nahvi A, Lee M, Jona I, Breaker RR (2006) Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA (New York, N.Y.) 27(1): 607-619.
- Ferré-D'Amaré AR (2010) The glmS ribozyme: use of a small molecule coenzyme by a gene-regulatory RNA. Q Rev Biophys 43(4): 423-447.
- Ferré-D'Amaré AR (2013) Riboswitches, in Encyclopedia of Biological Chemistry 2nd (Edn). Academic Press: Waltham pp. 136-141.
- Yu CH, Olsthoorn RCL (2015) Chapter Nineteen - Monitoring Ribosomal Frameshifting as a Platform to Screen Anti-Riboswitch Drug Candidates, in Methods in Enzymology. Academic Press pp. 385-393.
- Machtel P, Bąkowska-Żywicka K, Żywicki M (2016) Emerging applications of riboswitches - from antibacterial targets to molecular tools. Journal of Applied Genetics 57(4): 531-541.
- Winkler WC, Ali Nahvi, Adam Roth, Jennifer A Collins, Ronald R Breaker (2004) Control of gene expression by a natural metabolite-responsive ribozyme, Nature 428(6980): 281-286.
- Hampel KJ, Tinsley MM (2006) Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry 45(25): 7861-7871.
- Jansen JA, McCarthy TJ, Soukup GA, Soukup JK (2006) Backbone and nucleobase contacts to glucosamine-6-phosphate in the glmS ribozyme. Nat Struct Mol Biol 13(6): 517-23.
- McCarthy TJ, Plog MA, Floy SA, Jansen JA, Soukup JK, et al. (2005) Ligand requirements for glmS ribozyme self-cleavage. Chem Biol 12(11): 1221-1226.
- Mayer G, Famulok M (2006) High-throughput-compatible assay for glmS riboswitch metabolite dependence. Chembiochem 7(4): 602-604.
- Blount K, Puskarz I, Penchovsky R, Breaker R (2006) Development and application of a high-throughput assay for glmS riboswitch activators. RNA Biol 3(2): 77-81.
- Klein DJ, Ferré D, Amaré AR (2006) Structural Basis of glmS Ribozyme Activation by Glucosamine-6-Phosphate Science. Jstor 313(5794): 1752.
- Schüller A, Matzner D, Lünse CE, Wittmann V, Schumacher C, et al. (2017) Activation of the glmS Ribozyme Confers Bacterial Growth Inhibition. Chem Bio Chem 18(5): 435-440.
- Matzner D, Schüller A, Seitz T, Wittmann V, Mayer G (2017) Fluoro-Carba-Sugars are Glycomimetic Activators of the glmS Ribozyme. Chemistry - A European Journal 23(51): 12604-12612.
- Valsamatzi-Panagiotou A, Popova K, Penchovsky R (2020) Drug Discovery for Targeting Drug Resistant Bacteria. Springer 46: 205-228.

 Mini Review
Mini Review