Abstract
The aim of this work was to clone the full-length coding sequence of porcine DIS3L2 (DIS3 mitotic control-like protein) gene, search for polymorphism and perform association analyses between a gene-tagged SNP and litter size traits in the Large White and Landrace pig breeds. The full-length cDNA sequence of porcine DIS3L2 gene was cloned through the rapid amplification of cDNA ends (RACE) method. Sequence analysis revealed that the pig DIS3L2 gene encodes a protein of 883 amino acids which has high homology with the DIS3L2 protein of seven species: cattle (92%), giant panda (92%), horse (91%), dog (91%), human (89%), rat (85%) and mouse (85%). This novel porcine gene was assigned to GeneID: 100153005. This gene is structured in 20 exons and 19 introns as revealed by computer-assisted analysis. Phylogenetic analysis indicated that the pig DIS3L2 gene has a closer genetic relationship with the DIS3L2 gene of cattle. PCR– RFLP was established to detect the GU373702:c.1999C>A substitution of porcine DIS3L2 gene mRNA and association of this mutation with litter size traits was assessed in Large White (n = 200) and Landrace (n = 200) pig populations. Results demonstrated that this polymorphic locus was significantly associated with the litter size of all parities (P<0.01) in Large White and Landrace sows. Therefore, DIS3L2 gene is also a reproduction related gene.
Keywords: Pig, DIS3L2; Clone; Polymorphism; Peproduction
Introduction
DIS3 mitotic control-like protein (DIS3L2) is similar in sequence to 3’/5’ exonucleolytic subunits of the RNA exosome. The exosome is a large multimeric ribonucleotide complex responsible for degrading various RNA substrates. DIS3L2 is a tumor related gene for prior experiment indicated that DIS3L2 overexpression suppressed the growth of human cancer cell lines, and knockdown enhanced the growth of these cells. Germline mutations in DIS3L2 also cause the Perlman syndrome of [1]. DIS3L2 is an important development and reproduction related gene for it has been identified to be associated with the adult human height and mouse embryo development [2-5]. However, latest studies have shown that DIS3L2 gene has more important functions such as biological processes and metabolic processes of RNA binding, hydrolase activity, exonuclease activity, nuclease activity, molecular function, ribonuclease activity [6,7]. As mentioned above, DIS3L2 gene is an important gene which has many biological functions. Until today, DIS3L2 gene has been reported in human [8], cattle [5] and other animals, but, the swine DIS3L2 gene has not been reported yet.
The aim of this work was to clone the full-length porcine DIS3L2 mRNA sequence, search for polymorphism within this gene and perform association analyses between a gene-tagged SNP and litter size traits in the Large White and Landrace pig breeds.
Material and Methods
Ethics Statement
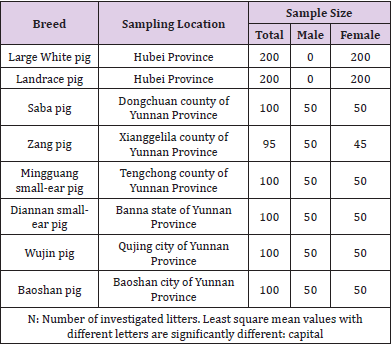
All procedures involving slaughtering, transport, or invasive procedures on live animals followed the guidelines for the care and use of experimental animals of the State Council (People’s Republic of China). This study was specifically approved by the ethics committee of Yunnan Agricultural University. Animals and Sample Preparation Six Large White pigs were slaughtered. Large instine, spleen, lung, muscle, fat, liver, heart, kidney and ovary samples were collected and snap frozen in liquid nitrogen then stored at – 80 °C. The total RNA was extracted using the Total RNA Extraction Kit (Gibco, USA). These RNA samples were used to perform RACE PCR. Ear samples were collected from 995 unrelated animals belonging to eight swine populations presented in Table 1. Genomic DNA isolated from these ear samples would be used to perform the polymorphism analyses. Both the total number of piglets born (TNB) and the number of piglets born alive (NBA) of 200 Large White sows and 200 Landrace sows listed in Table 1 were recorded for 700 litters. The litter size traits data and genomic DNA of these pigs would be used to perform association analyses.
5’- and 3’-RACE
5’- and 3’-RACE were performed to isolate the full-length cDNAs for pig DIS3L2 gene as the instructions of BD SMART™ RACE cDNA Amplification Kit (BD science, USA). For the pig DIS3L2 gene, the Gene-Specific Primers (GSPs) were designed based on pig EST sequence whose sequence is highly homologous to the coding sequence of human DIS3L2 gene: FD611611. 5’-RACE GSP: 5’- GCTTTCGATCATGCTCTGTGCATGT -3’, 3’-RACE GSP: 5’- CGGACCACCGAGTGCCTAGAATTTA -3’. RACE touchdown PCRs were carried out with 5 cycles of 94°C / 30 s and 72°C /3 min, followed by 5 cycles of 94°C / 30 s, 70°C / 30 s and 72°C / 3 min, finally with 30 cycles of 94°C / 30 s, 67 °C / 30 s, 72°C / 3 min to terminate reaction. The RACE PCR products were then cloned into pMD18-T vector (TaKaRa, Dalian, China) and sequenced bidirectionally with the commercial fluorometric method (SHENGGONG, Shanghai, China). At least five independent clones were sequenced for each PCR product.
Sequence Analysis
The gene analysis was conducted by GenScan software (http:// genes.mit.edu/GENSCAN.html). The protein comparison was performed using the Tool of BLAST at the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm. nih.gov/BLAST) and the ClustalW software (http://align.genome. jp/). The theoretical isoelectric point (pI) and molecular weight (Mw) of proteins was computed using the Compute pI/Mw Tool (http://www.expasy.org/tools/pi_tool.html).
PCR-RFLP
The DNA from the above pigs Table 1 was used as plate to perform PCR with primers: 5’- TGA CCT TTC CTC CCC AGA -3’(forward primer2), 5’- TGC TGG CTG CCA GAG ATA -3’ (reverse primer2). The PCR product size is 404bp. The 25 μL reaction system was: 2.0 μLDNA (100 ng), 2.5 μL 2 mM mixed dNTPs, 2.5 μL 10× Taq DNA polymerase buffer, 2.5 μL 25 mM MgCl2,1.0 μL 20 μM forward primer, 1.0 μL 20 μM reverse primer, 1.0 units of Taq DNA polymerase (1U /1 μL) (JINMEI, BIOTECH), and 12.5 μL sterile water. PCR was run as follows: 94°C for 4 min, followed by 35cycles of 94°C for 50 s, 57°C for 50 s, 72°C for 1 min, then 72°C extension for 10min, finally 4°C to terminate reaction. The 31 μL PCR-RFLP reaction volume was: 10 μL PCR product, 18 μL sterile water, 1 μL Taq I (10 U), 2 μL10 × buffer. The mixture was incubated in an air incubator at 65°C for 4 h, and then the genotypes were analyzed on the agarose gel (2.5%) containing ethidium bromide.
Statistical Analysis
The relationships between DIS3L2 genotypes and litter size traits of Large White (n=200) and Landrace (n=200) sows were evaluated with the general linear model (GLM) procedure of SAS version 8.0. Both additive and dominance effects were also estimated using REG procedure, where the additive effect was estimated as -1, 0 and 1 for AA, CA and CC genotype, respectively; and the dominance effect represented as 1, -1 and 1 for AA, CA and CC genotype, respectively [9]. The model: Yijkl =μ+ Pi +Sj + Fk+ Gl + eijkl, where Yijkl is the observation of the trait, μ is the least square means, Pi is the effect of ith parity (i =1, 2, 3, 4,5,6,7 (parity ≥ 7)), Sj is the effect of jth season, Fk is the effect of kth farm (k=1, 2), Gl is the effect of lth genotype (l = 1-3) and eijkl is the random residual [10].
Results
RACE Results for Pig DIS3L2 Gene
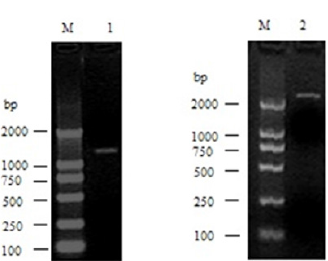
For pig DIS3L2 gene, through 5’-RACE, one PCR product of 1656bp was obtained. The 3’-RACE product was 2214 bp. These products were then cloned to T-vector and sequenced. These products were then cloned to T-vector and sequenced. Taken together, a 3173-bp cDNA complete sequence was finally obtained (Figure 1).
Figure 1: RACE results for pig DIS3L2 gene. M, DL2000 DNA markers 1, 5’-RACE product for pig DIS3L2 gene; 2, 3’-RACE product for pig DIS3L2 gene.
Sequence Analysis
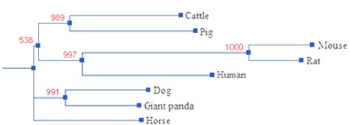
The cDNA nucleotide sequence analysis using the BLAST software revealed that this gene was not homologous to any of the known pig genes and it was then deposited into the GenBank database (Accession number: GU373702). The sequence prediction was carried out using the GenScan software and results showed that the 3173-bp cDNA sequence represented one single gene which encoded 883 amino acids. The theoretical isoelectric point (pI) and molecular weight (Mw) of these deduced proteins of this pig gene were computed using the Compute pI/Mw Tool. The pI of pig DIS3L2 is 6.09. The molecular weight of this putative protein is 99217.32. This novel porcine gene was assigned to GeneID: 100153005. BLAST analysis revealed that the pig DIS3L2 protein has high homology with the DIS3 mitotic control-like protein (DIS3L2) of seven species: cattle (accession number: NP_001179435, 92%), horse (accession number: XP_001495438, 91%), giant panda (accession number: XP_002918016; 92%), dog (accession number: XP_852722; 91%), human (accession number: NP_689596, 89%), rat (accession number: NP_001165630, 85%), mouse (accession number: NP_705758, 85%). Based on the results of the alignment of DIS3L2 proteins, phylogenetic trees were constructed using the Dendrogram procedure of ClustalW software (http://align.genome. jp/), as shown in (Figure 2).
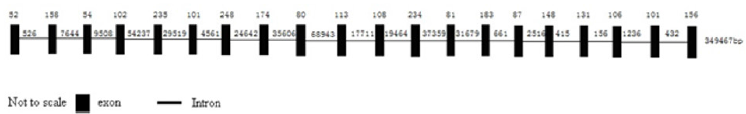
Phylogenetic analysis revealed that the pig DIS3L2 gene has a closer genetic relationship with the DIS3L2 gene of cattle. To obtain the genomic DNA of DIS3L2, the publicly available pig genome database at the NCBI Pig Genome Resources (http://www.ncbi.nlm. nih.gov/projects/genome/guide/pig/) was screened using the fulllength cDNA sequence of DIS3L2 as a seed. Sus scrofa chromosome 15(GenBank accession no. NC_010457) which encompasses entire DIS3L2 gene was identified by BLASTGen analysis. The pig DIS3L2 gene is 349467 bp in length and consists of 20 exons and 19 introns. All exon–intron splice junction sequences conform to the GT–AG rule (Figure 3).
Polymorphism
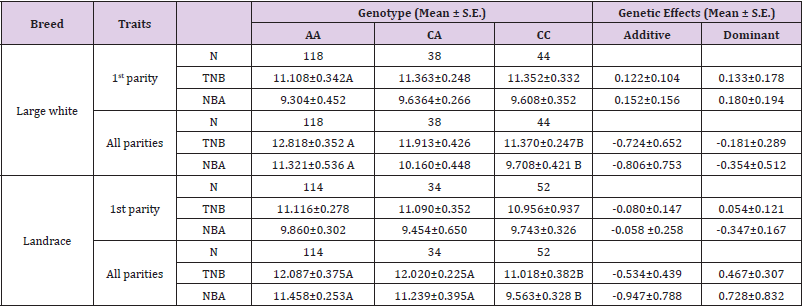
Based on the sequence of pig DIS3L2 gene, primers (forward primer2 and reverse primer2) were used to amplify the DNA of Large white and Landrace and the products were then cloned into PMD18-T vector and sequenced bidirectionally with the commercial fluorometric method. One C-A mutation was found at the position of 1999-bp of the coding region. This leaded to mutation of one Taq I restriction site. This was confirmed by PCR-Taq I-RFLP (Figure 4). Subsequently, PCR-Taq I-RFLP were performed using the DNA from 995 unrelated animals belonging to eight pig populations including Large White pig, Landrace pig, Saba pig, Zang pig, Mingguang small-ear pig, Diannan small-ear pig, Wujin pig and Baoshan pig Table 2. Results revealed that the frequency of A allele in the two exotic pig breeds: Large White pig (0.685) and Landrace pig (0.655) is higher than that in other six Yunnan local pig breeds: Saba pig (0.510), Zang pig (0.540), Mingguang small-ear pig (0.460), Diannan small-ear pig (0.460), Wujin pig (0.480) and Baoshan pig (0.495). The two exotic pig breeds: Large White pig and Landrace pig have more animals of genotype CA and AA .This indicated that Yunnan local pig breeds and exotic pig breeds displayed obvious genotype and allele frequency differences at this C-A mutation locus. Association of the SNP and litter size was assessed in two populations [purebred Large White and purebred Landrace sows] Table 3. Statistical analysis demonstrated that, for the first parity, no significant difference was found between animals of the three genotypes in the experimental purebred Large White sows and in the experimental purebred Landrace sows. For all parities, in the purebred Large White sows, those with the AA genotype also had an additional 1.448 piglets born compared to the CC animals (P<0.01) and 1.613 piglets born live compared to the CC animals (P<0.01). In addition, for all parities, in the purebred Landrace sows, AA and CA animals had 1.069 and 1.002 more piglets born than the CC animals, respectively (P<0.01), and 1.895 and 1.676 more piglets born alive than the CC animals, respectively (P<0.01).
Figure 4: The polymorphism analysis on pig DIS3L2 gene by PCR-Taq I-RFLP. CC: 404bp; CA: 404bp+228bp+176bp; AA: 228bp+176bp.
Discussion
With the development of modern bioinformatics and specific pig NCBI EST database was established along with different convenient analysis tools make researchers much easier to find the useful ESTs which was highly homologous to the coding sequence of human genes. Based on these swine EST sequences, we can obtain the complete coding sequences of some novel pig genes through the some modern experimental methods such as rapid amplification of cDNA ends (RACE) method. From the clone and sequence analysis of pig DIS3L2 gene, it could be seen that this is an effective method to isolate some novel pig genes. Through sequence analysis, it can be seen that the encoding protein of swine DIS3L2 gene is highly homologous with DIS3L2 proteins of human, cattle and other mammals. This implied that the DIS3L2 genes were highly conserved in some mammals and the swine DIS3L2 gene might have similar functions as the DIS3L2 genes of human, mouse and other mammals. We also found that the swine DIS3L2 protein does not show complete identity to that of human, cattle and other mammals. This suggested that the swine DIS3L2 gene might have some differences in functions to the DIS3L2 genes of human, cattle and other mammals.
Involvement of DIS3L2 gene in the mouse embryo development was a molecular basis for association analysis of this gene DNA polymorphism with litter size traits of pig [10-12]. Based on association analysis, it can be seen that the polymorphism (GU373702:c.1999C>A) of porcine DIS3L2 gene can significantly affect litter size. The AA genotype animals obviously have better litter size of all parities than the CC animals both in purebred Large White and purebred Landrace sows. This indicates that this polymorphic locus of porcine DIS3L2 gene is a valuable marker deserved to be applied to the marker assistant selection (MAS) in pig breeding. Therefore, DIS3L2 gene could be an useful candidate gene in selection for increasing litter size in pigs.
Conclusions
In conclusion, we first isolated the pig DIS3L2 gene and performed necessary sequence analysis, polymorphic analysis and association analysis. Our results suggest that DIS3L2 is not only a tumor related gene but also a fertility related gene. This established the primary foundation for further insight into this novel pig gene.
Acknowledgements
This work was supported by the Open Project of Yunnan Key Laboratory of Fertility Regulation and Minority Eugenics (2014– 2017), the Program for Innovative Research Team (in Science and Technology) in the University of Yunnan Province (No. 2014-22) and the National Nature Science Foundation of China (No. 31460645).
References
- Astuti D, Morris MR, Cooper WN, Staals RH, Wake NC, et al. (2012) Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumorsusceptibility. Nat Genet 44(3): 277-284.
- Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, et al. (2008) Many sequence variants affecting diversity of adult human height. Nat Genet 40(5): 609-615.
- Estrada K, Krawczak M, Schreiber S, van Duijn K, Stolk L, (2009) A genome-wide association study of northwestern Europeans involves the C-type natriuretic peptide signaling pathway in the etiology of human height variation. Hum Mol Genet 18(18): 3516-3524.
- Kimura K, WDiez-Roux G, Banfi S, Sultan M, Geffers L, et al. (2011) A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol 9(1): e1000582.
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309(5740): 1564-1566.
- Watahiki A, Waki K, Hayatsu N, Shiraki T, Kondo S, et al. (2004) Libraries enriched for alternatively spliced exons reveal splicing patterns in melanocytes and melanomas. Nat Methods 1: 233-239.
- Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, et al. (2009) A whole-genome assembly of the domestic cow, Bos Taurus. Genome Biol 10 R42.
- Akamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, et al. (2006) Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res16: 55-65.
- Niu BY, Ye LZ, Li FE, Deng CY, Jiang SW, et al. (2009) Identification of polymorphism and association analysis with reproductive traits in the porcine RNF4 gene. Anim Reprod Sci 110(3-4): 283-292.
- Zhang YH, Mei SQ, Peng XW, Niu BY, Ren ZQ, et al. (2009) Molecular characterization and SNPs analysis of the porcine Deleted in AZoospermia Like (pDAZL) gene. Anim Reprod Sci 112(3-4): 415-422.
- Morris MR, Astuti D, Maher ER (2013) Perlman syndrome: overgrowth, Wilms tumor predisposition and DIS3L2. Am J Med Genet C Semin Med. Genet 163(2): 106-113.
- Y Kamatani Y, Takahashi A, Matsuda K. Hosono N, Ohmiya H, et al. (2010) A genome-wide association study in 19 633 Japanese subjects identified LHX3-QSOX2 and IGF1 as adult height loci. Hum Mol Genet 19: 2303-2012.

 Research Article
Research Article