Abstract
Aim: Serratia marcescens is pathogen associated with nosocomial outbreaks among immunocompromised individuals. Molecular typing methods are useful tools for determining genetic relationships among bacterial isolates. The present study reports the analysis of the clonal relationships among S. marcescens isolates from an outbreak occurred in the neonatal unit (NU) at a referral public hospital of Belém, Pará, Brazilian Amazon, using PFGE and rep-PCR.
Material and Methods: Thirty isolates were obtained from referral hospital in the Brazilian Amazon. The molecular typing of isolates was performed using pulsed field gel electrophoresis (PFGE) and semiautomated rep-PCR in the DiversiLab System®.
Results: Results revealed that by PFGE there was the formation of three main clusters containing three, 13 and nine isolates. The DiversiLab rep-PCR analysis identified an agreement of 80.64% (25 of the 31 strains) compared to PFGE.
Conclusions: The semiautomated rep-PCR was similarly efficient as the PFGE for S. marcescens, demonstrating to be as efficient tool for outbreaks investigation.
Introduction
Serratia marcescens is microorganism often associated with nosocomial outbreaks and opportunistic infections including, bloodstream infections, urinary tract infections, central nervous system infection and pneumonias [1]. Episodes of colonization and outbreaks caused by this emergent pathogen are more likely to occur in Neonatal Intensive Care Units (NICU), with overall mortality and mobidity rates up to 58% [2-4]. The investigation of S. marcescens spread within nosocomial settings can be achieved by several molecular typing methods that assesses the genetic relatedness of isolates. Strain typing is relevant to determine if epidemiologically related isolates are also genetically related, establishing clonality of pathogens, and allowing the identification of infection sources [5,6]. The Pulsed Field Gel Electrophoresis (PFGE) is considered the reference fingerprinting approach for molecular epidemiology and identification of clonally-related strains of several bacterial pathogens [5-7]. Despite its high genetic discriminatory power, characteristics such as requirement of specialized equipment, challenging standardization, time and technically demanding nature could hinder its application in the occurrence of an outbreak, where investigation must be rapid and reliable [8,9].
Semiautomated system for strain typing based on the rep-PCR – the DiversiLab Microbial Typing System is based on amplification of regions between non-coding repetitive sequences in bacterial genomes [10]. The rep-PCR on the DiversiLab System provides rapid bacterial typing, simple standardization, reduced time execution and better comparison between results, being a satisfactory alternative to PFGE for evaluation of nosocomial outbreaks as demonstrated on previous studies involving healthcare-associated pathogens like Acinetobacter baumannii, Pseudomonas aeruginosa and Enterococcus faecium [9,11,12]. Outbreak investigations based on rep-PCR in different settings and populations are essential to confirm it as a molecular approach that may be used alone, also there is a limited number of reports analyzing its performance in comparison to PFGE and its discriminatory power. The present study reports the analysis of the clonal relationships among S. marcescens isolates from an outbreak occurred in the Neonatal Unit (NU) at a referral public hospital of Belém, Pará, Brazilian Amazon, using PFGE and rep-PCR.
Materials and Methods
Study Setting and Samples
A total of 30 S. marcescens isolates were obtained during an outbreak occurred between February and June 2006 in the NU of a referral hospital in Belém, Pará, Brazil. The unit is comprised of an Internal Nursery (IN), a Neonatal Intensive Care Unit (NICU) and an External Nursery (EN). The IN is divided into five wards: Special Care (SC), Intermediate Care (IC), Semi-Intensive (SI), other disorders and the transition room. The isolates were recovered from blood and swab rectal of neonates, hands of healthcare workers and environment. One isolate from other unit epidemiologically unrelated to the outbreak was included as a control.
Bacterial Isolates Identification
Isolates recovered from blood were cultured in the automated system BACTEC 9,120 (Becton Dickinson) and the others were plated onto sheep blood agar (Difco), Cled agar (Difco) and Mac-Conckey agar (Difco) and incubated at 37°C for 24h. Bacterial identification was performed using a Vitek® 2 microbial identification system (bioMérieux, Marcy l’Etoile, France).
Molecular Typing by PFGE
PFGE typing was performed out according to the Gautom et al. [13], with modifications. The DNA was digested on agarose miniblocks using the restriction enzyme XbaI at 37°C for 16 to 18 hours, and the fragments were separated by electrophoresis with a CHEF-DR II System (Bio-Rad Laboratories) for 24 hours at 160 V with the pulse initial of 5s and final 60s in Tris-borate-EDTA running buffer 0,5X. The DNA banding profiles were analyzed using the BioNumerics 6.5 software (Applied Maths, Belgium), with the similarity calculated based on the Dice coefficient (3% of tolerance).
Molecular Typing by Semi-Automated rep-PCR
Genomic DNA was extracted from a 10-μl loopful of a S. marcescens colony using the UltraClean Microbial DNA isolation kit (MoBio Laboratories, Carlsbad, CA), according to the manufacturer’s instructions. The extracted DNA was quantified on NanoDrop®2100c spectrophotometer (Thermo Scientific, USA) and adjusted concentration between 25ng/μl and 50ng/μl for each isolate were used for molecular essays. DNA was amplified using the DiversiLab Serratia kit (bioMérieux, France), following the manufacturers’ recommendations, together with template DNA. Thermal cycling conditions were initial denaturation at 94°C for 120 s, 35 cycles of denaturation at 94°C for 30 s, annealing at 65°C for 30 s, extension at 70°C for 90 s, and extension at 70°C for 180 s. The rep-PCR products were obtained after electrophoresis on the Agilent 2100 Bioanalyzer (Agilent Technologies, GER) using a microfluidics LabChip and analyzed on the Diversilab System® platform.
Results
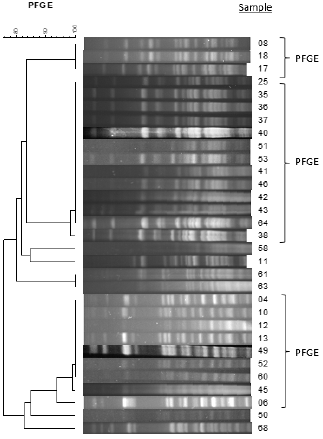
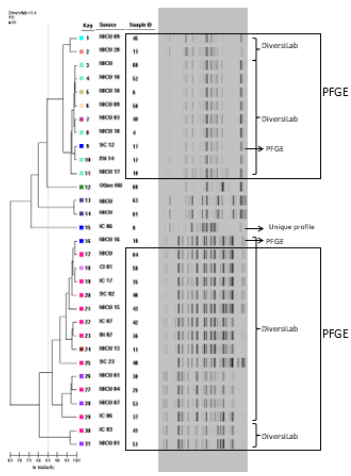
PFGE results revealed the presence of three main clusters (I, II and III) containing three, 13 and nine isolates respectively (Figure 1). For semiautomated rep-PCR on the Diversilab System® a similarity index of 85%, that provided the best rep-PCR agreement in comparison with PFGE, was applied [14]. The semiautomated rep- PCR on the Diversilab demonstrated an agreement of 80.64% (25 of the 31 strains) compared to PFGE. Three isolates assigned to PFGE I cluster were separated, two of these were included in the rep-PCR clusters IIa and IIIb, and one defined as a unique profile (ID 8). Three other strains, unique patterns by PFGE, grouped into clusters IIa (strains ID 58 and ID11) and IIIb (strain ID50). The cluster II were divided into two subgroups, IIa and IIb (80.5% similar). The cluster III revealed a higher diversification with two subgroups (IIIa and IIIb) with similarity above 93% similar (Figure 2).
Figure 2: DiversiLab generated dendrogram representing the rep-PCR fingerprint patterns for the 31 S. marcescens isolates. A similarity indice of 85% was used for interpretation.
Discussion
Although, considered the “gold standard” for epidemiologic studies because of its high discriminatory power, PFGE is timeconsuming. The Semiautomated rep-PCR in the DiversiLab System® has been proved to be a rapid and reliable method for molecular analysis of suspected nosocomial outbreaks caused by different microorganisms [9,15-18]. In the present study, the Semiautomated rep-PCR in the DiversiLab provided results similar to those obtained by the PFGE typing method, and also demonstrated a higher genetic diversity. Strains belonging to subgroup III, which, when analyzed by the rep-PCR, was divided into IIIa and IIIb with similarity around 93%. This occurrence can be explained due to the fact that:
a) The DiversiLab System® do not only consider the presence or absence of bands when differentiate isolates, but also bands intensity, which can decrease the percentage of similarity between strains that have the fingerprint
b) Approach of each method, since the PFGE is based on genomic restriction, while the rep-PCR is based on amplification of repeated palindromic sequences
c) The software for PFGE analysis – Bionumerics – allows the selection of the fingerprint bands to be analyzed, leading to a subjective evaluation, while DiversiLab System® has automatic marking bands, which do not allow manually interference in the assembly of the dendrogram.
The shared clonality relationship among the strains of a neonate (ID 12) and a health care professional (ID 60) suggests that these professionals can act as a vehicle for transmission of bacteria between ICUs and patients [19]. Sometimes, the usual measures of hand washing and patient care are not sufficient for prevention outbreaks, therefore, it is necessary to investigate health professionals, parents and others people that may be colonized and who come into contact with the neonates and, if necessary, also be included in treatment in order to make possible the complete eradication of the outbreak [20]. Catão [21] had been detected high level of transmission of Staphylococcus aureus from a team of nursing for children, newborn, indicating a high risk to the health of these patients. In addition, transmission can occur by direct contact from person to person, or by cross infection through indirect contact, which reinforces the importance of using investigative methods molecular typing methods, to identify and eradicate infection sources and transmission routes.
According to Ligozzi [5], differences in discriminatory power between PFGE and the semiautomated rep-PCR are depend on the bacterial species. Some reports have shown that rep-PCR has a less discriminatory power than PFGE for S. aureus and Enterococci, for other hand, other studies suggest concordance in both methods for Acinetobacter and Escherichia coli [15,22-26]. The first study that assessed the reliability of the semiautomated rep-PCR in the DiversiLab System® in comparison with PFGE for S. marcescens, suggested in their results an apparent increase in the genetic variability when evaluating DiversiLab system results, indicating that not all of the samples of a PFGE cluster showed the same level of similarity. According to our data, a relevant concordance between both methods was observed, with the rep-PCR also demonstrating an increase in genetic variability, confirming previously reported data [5].
The semiautomated DiversiLab rep-PCR system constitute a compelling tool for strain typing of S. marcescens during outbreak investigations. Due to the speed of obtaining results and their correlation with the reference standard for epidemiological investigation, the PFGE, this approache can assist in the rapid introduction of infection control measures. This study suggests that use of the DiversiLab in cases of outbreaks is reliable. Furthermore, the performance time is reduced to few hours when compared to the reference technique, which it takes about three days to obtain the results. The costs of both are comparable, however, the establishment costs of the DiversiLab system® are higher, but if the system is inserted in a laboratory that already performs molecular essays and has access to a thermocycler, the costs can be reduced [14].
Conclusion
The semiautomated rep-PCR was similarly efficient as the PFGE for S. marcescens genotyping, also presenting the potential for use with a wide range of pathogenic microorganisms. Further studies are recommended in order to confirm its effectiveness and extract its full potential for assists in rapid epidemiological investigation of outbreaks.
Conflict of Interest
The authors declare no conflict of interest regarding the publication of this study.
Funding
This research was supported by funding from Fundação de Amparo à Pesquisa do Pará/Universidade do Estado do Pará (FAPESPA/UEPA) [Cooperation grant N°004/2019], Programa Institucional de Bolsas de Iniciação Científica/Instituto Evandro Chagas - Conselho Nacional de Desenvolvimento Científico e Tecnológico (PIBIC/IEC-CNPq) and Instituto Evandro Chagas/ Ministério da Saúde/Secretaria de Vigilância em Saúde (IEC/MS/ SVS).
References
- Mahlen SD (2011) Serratia infections: from military experiments to current practice. Clinical microbiology reviews 24(4): 755-791.
- Kim SB, Jeon YD, Kim JH, Kim JK, Ann HW et al. (2015) Risk factors for mortality in patients with Serratia marcescens bacteremia. Yonsei medical journal 56(2): 348-354.
- Leng P, Huang WL, He T, Wang YZ, Zhang HN (2015) Outbreak of Serratia marcescens postoperative infection traced to barbers and razors. The Journal of hospital infection 89(1): 46-50.
- Voelz A, Müller A, Gillen J, Le C, Dresbach T, et al. (2010) Outbreaks of Serratia marcescens in neonatal and pediatric intensive care units: clinical aspects, risk factors and management. Int J Hyg Environ Health 213(2): 79-87.
- Ligozzi M, Fontana R, Aldegheri M, Scalet G, Lo Cascio G (2010) Comparative Evaluation of an Automated Repetitive-Sequence-Based PCR Instrument versus Pulsed-Field Gel Electrophoresis in the Setting of a Serratia marcescens Nosocomial Infection Outbreak. J Clin Microbiol 48(5): 1690-1695.
- Singh A, Goering RV, Simjee S, Foley SL, Zervos MJ (2006) Application of molecular techniques to the study of hospital infection. Clin Microbiol 19(3): 512-530.
- Tenover FC, Arbeit RV, Goering RV, Mickelsen PA, Murray Be, et al. (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33(9): 2233-2239.
- Maâtallah M, Bakhrouf A, Habeeb MA, Turlej Rogacka A, Iversen A, et al. (2013) Four Genotyping Schemes for Phylogenetic Analysis of Pseudomonas aeruginosa: Comparison of Their Congruence with Multi-Locus Sequence Typing. PLoS ONE 8(12): e82069.
- Ratkai C, Peixe LV, Grosso F, Freitas AR, Antunes P, et al. (2010) Successful application of the DiversiLab repetitive-sequence-based PCR typing system for confirmation of the circulation of a multiresistant Pseudomonas aeruginosa clone in different hospital wards. Diagnostic Microbiology and Infectious Disease, 67(2): 202-206.
- Healy M, Huong J, Bittner T, Lising M, Frye S et al. (2005) Microbial DNA Typing by Automated Repetitive-Sequence-Based PCR. J Clin Microbiol 43(1): 199-207.
- Werner G, Fleige C, Neumann B, Bender JK, Layer F (2015) Evaluation of DiversiLab®, MLST and PFGE typing for discriminating clinical Enterococcus faecium isolates. Journal of microbiological methods 118: 81-84.
- Higgins PG, Janssen K, Fresen MM, Wisplinghoff H, Seifert H (2012) Molecular epidemiology of Acinetobacter baumannii bloodstream isolates obtained in the United States from 1995 to 2004 using rep-PCR and multilocus sequence typing. J Clin Microbiol 50(11): 3493-500.
- Gautom RK (1997) Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other Gram-negative organisms in 1 day. J Clin Microbiol 35(11): 2977-2980.
- Ross TL, Merz WG, Farkosh M, Carroll KC (2005) Comparison of an automated repetitive sequence-based PCR microbial typing system to pulsed-field gel electrophoresis for analysis of outbreaks of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 43(11): 5642-5627.
- Grisold AJ, Zarfel G, Hoenigl M, Krziwanek K, Feierl G (2010) Occurrence and genotyping using automated repetitive-sequence–based PCR of methicillin- resistant Staphylococcus aureus ST398 in Southeast Austria. Diagn Microbiol Infect Dis 66(2): 217-221
- Tenover FC, Gay EA, Frye S, Eells SJ, Healy M (2009) Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulse-field gel electrophoresis. J Clin Microbiol 47(8): 2452-2457.
- Doléans Jordheim A, Cournoyer B, Bergeron E, Croizé J, Salord H (2009) Reliability of Pseudomonas aeruginosa semi-automated rep-PCR genotyping in various epidemiological situations. Eur J Clin Microbiol Infect Dis 28(9): 1105-1111.
- Grisold AJ, Zarfel G, Strenger V, Feierl G, Leitner E (2010) Use of automated repetitive sequence based PCR for rapid laboratory conformation of nosocomial outbreaks. J Infect 60(1): 44-51.
- Tragante CR, Ceccon MEJR, Falcão MC, Seiti M, Sakita N (2008) Prevalência de sepse por bactérias Gram negativas produtoras de beta-lactamase de espectro estendido em Unidade de Cuidados Intensivos Neonatal. Rev Paul Pediatr 26(1): 59-63.
- Rahal JJ, Urban C, Horn D, Freeman K, Segal Maurer S (1998) Class restriction of cephalosporin use to control total cephalosporin resistance in nosocomial Klebsiella. JAMA 280(14): 1233-1237.
- Catão RMR, Belém LF, Silva PMF, Luiza FP, Nunes LE (2012) Avaliação da colonização nasal por Staphylococcus aureus em funcionários de um serviço de saúde em Campina Grande-PB. 7(1): 1983-4209.
- Shutt CK, Pounder JI, Page SR, Schaecher BJ, Woods GL (2005) Clinical evaluation of the DiversiLab microbial typing system using repetitive-sequence-based PCR for characterization of Staphylococcus aureus strains. J Clin Microbiol 43(3): 1187-1192.
- Pounder JI, Shutt CK, Schaecher BJ, Woods GL (2006) Clinical evaluation of repetitive sequence-based polymerase chain reaction using the DiversiLab System for strain typing of vancomycin-resistant enterococci. Diagn. Microbiol Infect Dis 54(3): 183-187.
- Carretto E, Barbarini D, Farina C, Grosini A, Nicoletti P (2008) Use of the DiversiLab semiautomated repetitive-sequence-based polymerase chain reaction for epidemiologic analysis on Acinetobacter baumannii isolates in different Italian hospitals. Diagn Microbiol Infect Dis 60(1): 1-7.
- Fontana C, Favaro M, Minelli S, Bossa MC, Testore GP (2008) Acinetobacter baumannii in intensive care unit: a novel system to study clonal relationship among the isolates. BMC Infect Dis 8: 79.
- Pitout JDD, Campbell L, Church DL, Wang PW, Guttman DS (2009) Using a commercial DiversiLab semiautomated repetitive sequence-based PCR typing technique for identification of Escherichia coli clone ST131 producing CTX-M-15. J Clin Microbiol 47(4): 1212-1215.

 Research Article
Research Article