Abstract
Objective: Hamamy syndrome is a recently discovered rare genetic disorder caused by a missense mutation of Iroquois (IRX5) gene. IRX-5 transcription factor orchestrates cell movement in the developing fetus which underlies head and gonad formation. It also modulates multiple organs during embryogenesis like heart, bone, blood, facial appearances. Study animals play an essential role in understanding the pathogenesis of diseases and in pre-clinical studies of drug development. Hence our study was aimed at conducting comparative genomic analysis to find suitable animal models with similar sequence for IRX-5 gene that can be used for further studies. In our preliminary study we used Bioinformatics software CLC sequence viewer 6.7 to study three parameters; multiple sequence alignment, phylogenetic relationships and protein analysis of the IRX-5 gene.
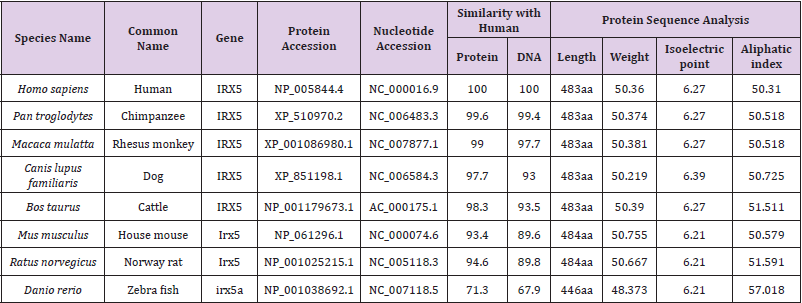
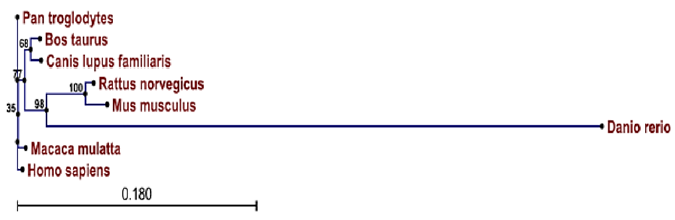
Results: Comparative genomic studies of the human IRX-5 showed high degree of homology with respect to IRX-5 of P. troglodytes(99.6%), M.mulatta(99%), B.taurus(98.3%), C. lupus(97.7%), M.musculus(93.4%), R.norvegicus(94.6) and lower homology to D.rerio(71.3%). These results further revealed a closer phylogenetic relationship of Homo sapiens IRX-5 with P. troglodytes, M.mulatta, B. Taurus, C. lupus, than with M.musculus, R.norvegicus and D.rerio.
Keywords: Comparative Genomics; Iroquois Gene; Hamamy Syndrome; Animal Model; Genome Correspondence; Gene Identification; Genome Evolution
Abbreviations: HGP- Human Genome Project; NCBI- National Center for Biotechnology Information; BLAST- Basic Local Alignment Search Tool; EURORDIS- European Organization for Rare Diseases; FDA- Food and Drug Administration Agency; MSAMultiple Sequence Alignment; OMIM- Online Mendelian Inheritance in Man; OD- Orphan Disease
Introduction
A rare disease, also referred to as an Orphan Disease (OD), is any disease that affects a small percentage of the population. To date 6000-8000 rare diseases have been identified worldwide, and their number is increasing as a result of improved diagnostic techniques. A rare disease is defined as disease affecting less than 200,000 people in the United States, and less than 500 people per 1 million in Europe according to the European Organization for Rare Diseases (EURORDIS). Many rare diseases appear early in life and it has been reported that 75% of all patients with rare diseases are children and 30% of the children with rare diseases die within 5 years after birth. In the developed countries 10% of the population has some form of ODs. [1-5] EURORDIS estimated that at least 80% of rare diseases are identified with genetic origins have little or no treatment options, not rigorously studied and thus are present in person’s entire life, even if symptoms do not immediately appear [6].
Hamamy syndrome is a rare autosomal recessive human syndrome 5 (MIM611174) was described by Hamamy et al first in two Jordanian brothers born to double first cousin parents in 2007 [7] later in Turkey with two brothers born to a consanguineous marriage [8-9]. Hamamy syndrome is caused by a missense mutation of the IRX5(Iroquois) gene which results in the formation of defective proteins that are necessary for the morphogenesis of the heart, brain, face, bone and gonads. It also contributes to the morphogenesis of secondary organs such as dental, lachrymal, salivary and thyroid structures [10]. The Iroquois (IRX) gene is a homeobox gene i.e., class of gene involved in the regulatory functions of embryonic development [6-8] and encodes six transcription factors which are conserved across species. The six IRX genes have been organized into two-three gene complexes i.e., IRXA which contains IRX-1, IRX-2, IRX-4 genes and IRXB which contains IRX- 3, IRX-5 and IRX-6 genes [11]. The IRX-5 gene located on the long arm of chromosome 16 is involved in modulating the migration of progenitor cells in the branchial arches and gonads [8].
Clinically, Hamamy Syndrome is characterized by craniofacial dysmorphisms, including severe tele canthus, myopia, hypoplasia of the lacrimal-salivary apparatus, frontonasal anomalies, protruding ears and sensory-neural hearing impairment. Skeletal anomalies with repeated fractures of long bones, hypodontia, microcytic hypochromic anemia and congenital heart defects with intraventricular conduction delay have also been documented in all affected individuals [9]. To encourage studies focusing on the development of products to treat ODs, the Food and Drug Administration Agency (FDA) passed the 1983 Orphan Drug Act. Within the first 25 years of passing this act in the US, 326 products which target more than 200 rare diseases have been approved. Although this is a good start, until now therapy is available for only 5% of all the known ODs. The small market size and thus revenue of ODs, lack of a suitable animal model, lack of adequate clinical data, unknown or unclear etiology, problems in finding sufficient number of patients with the disease and scarcity of funding have been some of the problems cited into the lack of progress in the development of therapy for ODs [1-4].
Main Text
Methodology
Bioinformatics Tools Used: BLAST (Basic Local Alignment Search Tool) of NCBI (National Centre for Biotechnology Information), HomoloGene, UniGene, NCBI Map Viewer and CLC Sequence Viewer of Version 6.7.
Methods
1. Homo sapiens IRX-5 sequence similarity search:
Protein sequences of IRX-5which closely matches with Homo sapiens IRX-5were identified using BLAST tool supported through NCBI, USA. BLAST tool is designed to identify potential homologues for a given sequence using a set of algorithms can be used to analyse both nucleotide and protein sequence of interest. Identification of homologues will help in the prediction of unidentified functions of proteins or genes, for identifying species, locating domains, establishing phylogeny, DNA mapping and also in modelling of the 3-D structure of new proteins. There are different types of BLAST programs available and each of them serves for specific purpose [15].
2. Retrieval of IRX-5 protein sequences:
After sequence similarity search from the BLAST tool, the complete protein sequences of IRX-5 of other species which showed close match to IRX-5of Homo sapiens were retrieved from NCBI database and used for further comparative study.
3. In Silico comparison of Homo sapiens IRX-5 with other species:
All the retrieved IRX-5 sequences of other species were compared with Homo sapiens IRX-5 using bioinformatics software CLC Sequence Viewer Version 6.7. The parameters studied for comparison were multiple sequence alignment, Phylogeny and Protein statistics.
a. Multiple Sequence Alignment:
Multiple sequence alignment is a process of arranging the sequences of nucleotides or protein to identify the areas of similarity between the compared sequences which may occur as a result of functional, structural, or evolutionary relationships. Sequence alignment of nucleotide or amino acid residues are usually presented as rows, so that aligned sequences will appear in successive columns. Whereas in case of text formats, aligned columns containing identical or similar characters are designated with specific conservation symbols. For protein sequence alignments, color sare often used to indicate amino acid properties to assist in deciding the conservation of a given amino acid substitution. In multiple sequence alignment, the last row in each column often represents the consensus sequence. In some cases, consensus sequence is also represented in graphical format with a sequence logo, where the magnitude of each nucleotide or amino acid letter relates to its extent of conservation.
b. Phylogeny: [16]
The ancestral relationships between living organisms can be denoted by using an evolutionary tree i.e., the tree of life. The tree of life characterizes the phylogeny of organisms i.e., the description of organismal roots as they change through time. By exploring the phylogeny of organisms, we can easily find out any differences that exists among plants, animals and microorganisms. Phylogenetic and sequence alignment are interrelated fields due to the need of evaluating sequence relatedness. Phylogenetic study makes extensive use of sequence alignments in building and understanding of phylogenetic trees, which further used to determine the evolutionary relationships between the homologous genes presented in the genomes of divergent species. The degree to which sequences in a question set to differ is qualitatively linked to the sequence evolutionary distance from one another. For instance, high sequence identity indicates that the sequences in question are comparatively young and also have most recent common ancestor, while low identity suggests that the separation is more ancient.
Results and Discussion
In current scenario of medical research, comparative genomics applications has improved the outcome of genetic maps in human and other model organisms and detected many rare human disease genes in other mammalian models [17]. Sequence similarity search using Homolo Gene search tool found seven different species with IRX-5 gene having close similarity to Human IRX-5. The gene accession number, Pairwise Alignment Scores, and the complete IRX-5 Protein sequence statistics (length, isoelectric point and aliphatic index) of all the species compared are given in Table 1.
Multiple Sequence Alignment (MSA)
The main goal of sequence alignment is to enable the researcher to ascertain whether compared sequences display sufficient similarity, so that an inference of homology is justified. Although these two terms i.e., similarity and homology are often used interchangeably, but in practice the terms are different i.e., similarity is an observable quantity that might be expressed as percent identity and on the other hand, homology refers to a conclusion drawn from these data that two genes share a common evolutionary history. Genes either are or are not homologous, there are no degrees for homology as there are for similarity. In our study, the observed homology was ranged from 99.6% to 71.3% between the protein sequences of the Human IRX-5 and other animals which includes chimpanzee, rhesus monkey, dog, cattle, mouse, rat and zebra fish which were obtained by pair wise alignment scores using HomoloGene Tool. The high degree of homology was found with chimpanzee (99.6) and then to rhesus monkey (99.0), as compared to cattle (98.3%), dog (97.7%), mouse (93.4%), rat (94.6%) and zebra fish (71.3%). Results of multiple sequence alignment were shown in Figure 1. Completely conserved regions were represented by purple color, aqua color was used for most conserved, yellow color for semi conservative region and less conserved where represented by dark red color. Conservation scores were also indicated as bar chart for each amino acid similarity at the bottom.
Phylogeny
The animals described in this study could be classified into the five major groupsi.e., Rodentia (mouse and rats), Primates (Human, Chimpanzee and Rhesus monkey), Artiodactyla (cattle), Carnivora (dog) and Cypriniformes (Zebra fish). Phylogenetic analysis showed that the chimpanzee and rhesus monkey IRX-5 protein sequences have greatest homology to the published Human IRX-5 sequences as compared to others. Our results also indicated the relationship that human IRX-5 with the other animals in the order of Primates (Chimpanzee and Rhesus monkey), Carnivora (dog), Artiodactyla (cattle), Rodentia (mouse and rats) and then with Cypriniformes (Zebra fish). Phylogenetic relationship among the species studied is shown in Figure 2.
Conclusion
Although much has been achieved in the field of medical research through the use of traditional methods of animal model finding, but their widespread use is limited due to some inherent faults that prevent its relevance to clinical medicine [18]. As it is very crucial to use animal models, because testing of experimental hypothesis directly in human subjects is not always possible due to various ethical or logistical reasons. But with the help of comparative genomics, it is feasible to predict suitable animal models with highest precision through the detection of relationship in terms of structural and functional aspects of human genome and also the pathophysiological aspects of human disease with other animal models available. Ultimately, comparative genomics is becoming one of the most promising scientific fields today, based on its application in various field of study, we anticipate more and more exciting technological advances and important biological discoveries in the near future.
Limitations of the Study
Commonly, most of the complex diseases are diagnosed only at late stages and as a result have poor prognoses. Whereas with the help of comparative genomics, clinicians may able to detect and diagnose a complex disease in early stage and assure positive outcomes like extension and improvement in quality of life through better therapy. Finally, the data presented in this work can provide base line information and this alone is not sufficient to make decision among the alternatives, because we used free version of software with limited options and not the commercial version. Hence, our future plan is to continue the study with the more detailed comparison of functional aspects of Iroquois gene to other species.
Ethics Approval and Consent to Participate
This preliminary study got Asmara College of Health Sciences (ACHS) Ethical committee clearance and also granted permission to participate in the study.
Consent for Publication
All the Authors have agreed to publish this article.
Availability of Data and Material
The data used in this study is available from the corresponding author upon reasonable request.
Competing Interests
None
Funding
Not Applicable
Authors’ contribution
NSB has contributed in research idea, research framework, reviewing and editing the paper, ND, NH and SH are involved data retrieval, analysis, interpretation, presentation and writing the article with appropriate citations. All authors have read and approved the manuscript for publication.
Acknowledgement
We kindly acknowledge Asmara College of Health Sciences (ACHS) Ethical committee for granting the ethical clearance on time and also the college administration for proving us internet resource to conduct our study.
References
- Pinnapureddy AR, Stayner C, Mc Ewan J, Baddeley O, Forman J, et al. (2015) Large animal models of rare genetic disorders: sheep as phenotypically relevant models of human genetic disease. Orphanet Journal of Rare Diseases.
- Putzeist M, Mantel-Teeuwisse AK, Gipsen-de Wied CC, Hoes AW, Leufkens HG, et al. (2013) Drug development for exceptionally rare metabolic diseases: challenging but not impossible. Orphanet Journal of Rare Diseases 8: 179.
- Molineris I, Ala U, Provero P, Di Cunto F (2013) Drug repositioning for orphan genetic diseases through Conserved Anticoexpressed Gene Clusters (CAGCs). BMC Bioinformatics 14: 288.
- Miyamoto BE, Kakkis ED (2011) The potential investment impact of improved access to accelerated approval on the development of treatments for low prevalence rare diseases. Orphanet Journal of Rare Diseases 6: 49.
- Kakkis ED, O’Donovan M, Cox G, Hayes M, Goodsaid F, et al. (2015) Recommendations for the development of rare disease drugs using the accelerated approval pathway and for qualifying biomarkers as primary endpoints. Orphanet Journal of Rare Diseases.
- Aymé S, Schmidtke J (2007) Networking for rare diseases: a necessity for Europe. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 50(12): 1477-1483.
- Hamamy HA, Teebi AS, Oudjhane K, Shegem NN, Ajlouni KM (2007) Severe hypertelorism, midface prominence, prominent/simple ears, severe myopia, borderline intelligence, and bone fragility in two brothers: new syndrome? American Journalof Medical Genetics 143A(3): 229-234.
- (2019) OMIM database.
- Bonnard C, Strobl AC, Shboul M, Lee H, Merriman B, et al. (2012) Mutations in IRX5 impair craniofacial development and germ cell migration via SDF1. Nature Genetics 44(6): 709-713.
- Hall BK (2008) The neural crest and neural crest cells: discovery and significance for theories of embryonic organization. Journal of Biosciences 33(5): 781-793.
- Kerner P, Ikmi A, Coen D, Vervoort M (2009) Evolutionary history of the iroquois/Irx genes in metazoans. BMC Evolutionary Biology.
- Harvey M (2007) Animal Genomics in Science, Social Science and Culture. Genomics, Society and Policy. 3(2): 1-28.
- Bezold KY, Karjalainen MK, Hallman M, Teramo K, Muglia LJ (2013) The genomics of preterm birth: from animal models to human studies. Genome Medicine 5(4): 34.
- Sivashankari S, Shanmughavel P (2007) Comparative genomics - A perspective. Bioinformation 1(9): 376-378.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J MolBiol 215(3): 403-410.
- Felsenstein J (2004) Inferring phylogenies. Sinauer associates, Inc: sunderland, Massachusetts.
- O'Brien SJ, Menotti-Raymond M, Murphy WJ, Nash WG, Wienberg J, et al. (1999) The promise of comparative genomics in mammals. Science 286(5439): 458-462, 479-481.
- Kelly SS, Meredith JM, Kapil V, Laurie AR, Jonathan EB, et al. (2004) Genomics and Clinical Medicine: Rationale for Creating and Effectively Evaluating Animal Models. Experimental Biology and Medicine 229(9): 866-875.

 Short Communication
Short Communication