Abstract
Objective: MecA gene sequence analysis was used as a tool for characterization of methicillin resistant Staphylococcus aureus. One hundred bacterial isolates were obtained from conjunctiva patients in different ophthalmic hospitals in Khartoum State. Samples were collected in enrichment selective media and biochemical tests were used to identify the bacteria as S. aureus. MRSA sensitivity were tested using the Kirby-Bauer disc diffusion method. DNA was extracted from those resistant S. aurei using modified guanidine thiocyanate (GTC) method. A universal primer was used to amplify MecA gene by a conventional PCR technique. The amplified PCR products from positive samples were sequenced, and the sequences were viewed by Finch TV program version 1.4.0. The identity and similarity of the nucleotide sequence of the isolated strains was detected by comparing them with published sequences using BLASTn.
Results: Results revealed that S. aureus were identified in (23.5%) patients. S. aureus isolates were found highly resistant to methicillin MRSA 89 (89.0%) and less resistant to vancomycin VRSA 6 (6.0%). Among 89 methicillin-resistant S. aureus (MRSA) and 11 methicillin-sensitive S. aureus (MSSA) strains 30/89 (33.7 %) were positive for mecA gene. Whereas all MSSA isolates were negative for the mecA gene. 5 samples (labelled 13, 17, 52, 58 and 140) were sent for characterization by sequencing of PCR products. The analysis of the mecA sequence revealed no obvious mutation in the gene. BLAST search at the Gen Bank database with the mecA sequences for other species of S. aureus displayed that were clearly closely related to MH807092.1, Sequence analysis by BLASTn displayed high similarity and identity with Staphylococcus aureus strain MecA-S1 from India MH807092 and with other Staphylococcus aureus strain MecA-S1 isolates from the Gen Bank database.
Conclusion: mecA gene PCR was found to be effective to validate the phenotypic detection of MRSA. This reveals the high specify of the primers and accuracy of the PCR. Thus, mecA sequencing can be used to identify genetically atypical MRSA isolates from different origins.
Keywords: MRSA; MecA gene; Sequence; PCR; Blast; Sudan
Introduction
Staphylococcus aureus comprises Gram-positive aerobic or facultatively anaerobic cocci in clusters, occurring as normal flora of the skin, axilla, and anterior nares of man and animals [1]. S. aureus cause minor skin infections, septicaemia, toxic shock, and pneumonia. The increasing incidence rate of methicillin was first described in 1961, and its spread in hospitals and the community, has posed a major challenge for infectious disease [2]. Bacterial conjunctivitis is the most widespread pattern of infective conjunctivitis [3]. The most common bacterial pathogen in conjunctivitis worldwide is due to Staphylococcus aureus [4]. Methicillin-resistant Staphylococcus aureus (MRSA) is considered as a leading global concern in the health sector and more recently in the community. The gene responsible for methicillin resistance is mecA, which is carried by a DNA fragment known as staphylococcal cassette chromosome mec (SCCmec). This encodes a protein called penicillin-binding protein (PBP-2a), which inhibits the action of β-lactam antibiotics such as methicillin [5]. The mecA gene complex contains insertion sites for other mobile genetic elements (e.g. plasmids and transposons) that facilitate the acquisition of resistance genes to other antibiotics. Epidemiological studies have shown that hospital- and community-acquired MRSA infections are increasing in many parts of the world [6].
Methicillin resistance is attributed to a modified penicillinbinding protein 2(PBP) of Staphylococcus aureus encoded by the mecA gene. This protein gives MRSA strains a reduced affinity for b-lactam antibiotics resulting in a normal cross-linking of peptidoglycan strands during cell wall synthesis [7]. The emergence and worldwide spread of methicillin resistant S. aureus strains is largely due to horizontal transfer of mobile genetic elements carrying resistance and virulence determinants [8,9-16]. This study aimed to highlight the importance of using sequencing technique in the validation of different suspected mutations with in the genome of S .aureus isolates.
Methods
Clinical Isolates
This was a cross-sectional, analytical study in which specimens were collected from Mecca Ophthalmic Hospitals, Khartoum Ophthalmic Hospital and Koch Ophthalmic Clinic (Khartoum State, Sudan). Conducting the research study was ethically approved by the Ethical Committee of the University of Science and Technology (Omdurman, Sudan). And informed consents were obtained from all study participants. Permission for specimen’s collection was granted by the authorities of the different hospitals and clinics mentioned above. A total of 426 eye swabs were collected from patients presenting with symptoms and signs of Conjunctivitis. All swabs were inoculated directly on blood agar (Hi-Media, India), MacConkey agar (Hi-Media, India) and chocolate agar, and all primary cultures were subculture on mannitol salt agar. One hundred S.aureus Isolates were identified by colonial morphology, Gram stain, and standard biochemical tests. Detection of MR was performed by disc diffusion method for S. aureus: Methicillin disc (5μg) (Hi-Media Company India) placed on Muller-Hinton agar with 5.0 per cent NaCl according to CLSI guidelines and incubated for 24 h at 35 ± 2ºC. MSSA (ATCC 6538) and MRSA (ATCC 33591) were included as positive controls. After overnight incubation, the diameter of each zone of inhibition was measured in mm. The susceptibility testing results were recorded according to the Clinical and Laboratory Standards Institute (CLSI) guidelines10. Interpretative criteria for disc diffusion method for S. aureus was as follow; resistant < 9mm, intermediate resistance 10- 13 mm and sensitive > 14mm. In order to distinguish strains exhibiting Intermediate resistant” from that of “heterogenous strains” the sensitivity plates with strains exhibiting resistance were incubated for an additional 24 hrs. At the end of 48 hrs of incubation, heterogenous strain turned sensitive whereas intermediate resistant strains remained resistant. All methicillin (oxacillin) resistant strains were identified and subjected to estimation of minimum inhibition concentration against oxacillin using the E-test method (Biomerieux, Marcy Etoile, France) as per manufacturer’s instructions.
DNA Extraction
DNA was extracted from subculture of a single colony after overnight incubation by modified guanidine thiocyanate (GTC) method11.
Conventional Polymerase Chain Reaction (PCR):
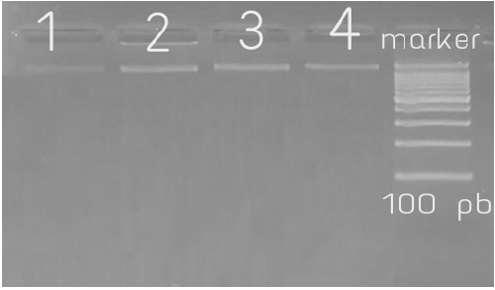
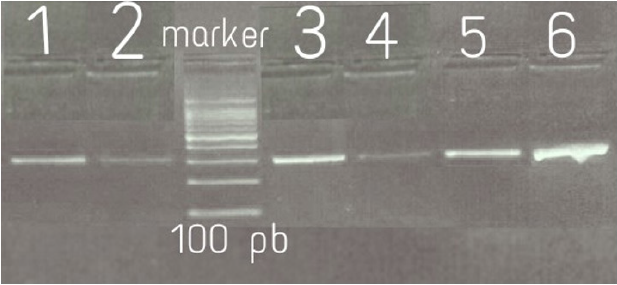
One hundred S.aureus genomic DNA samples were used as templates for PCR amplification of the 16S rRNA and MecA gene. The primers set used for16S rRNA were F (5’AGTTTGATCCTGGCTCAG3’) and R (5’AGGCCCGGGAACGTATTCAC3’). The set primers used for MecA gene were F(5’TGGCTATCGTGTCACAATCG3’) and R (5’CTGGAACTTGTTGAGCAGAG3’´) for forward primer and reverse primer, respectively (Macrogen, South Korea). The amplification was done using CLASSIC K960 China thermal cycler. DNA amplifies was done using Maxime PCR Premix kit (I-Taq) (iNtRON, Korea). The PCR assay was carried out in a total volume of 25μL of mixture containing 0.5 μl of each of the gene-specific primers (1 μl), 4μL of template DNA and 20 μl of WFI (water for injection). For amplification of 16S rRNA, oligonucleotide primers were used for Thermal cycling conditions as follows: initial denaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 40 sec, annealing 56°C for 40sec, extension 72°C for40 sec; final extension at 72°C for 5min. For amplification of mecA, oligonucleotide primers were used, the reaction conditions were initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 40 sec, primer annealing at 52°C for 45 sec, and extension at 72°C for 60 sec and final extension at 72°C for 2 min12. PCR products (5μl) were analyzed by gel electrophoresis in 2.0% agarose stained with ethidium bromide. The results were photographed under ultraviolet light machine (Transilluminater; Uvite, UK) to detect the specific amplified product by comparing it with 100 base pairs standard DNA ladder (Figures 1 & 2) and the remains from PCR products were store at -20°C until sequencing.
Figure 1: The 16s rRNA gene after PCR on 2% agarose gel: Lane M= 100 bp DNA Marker, lane 1, 2, 3and4 positive results for 16s rRNA of S.aureus (1500bp).
Figure 2: The mecA gene after PCR on 2%Agarose gel electrophoresis: Lane M= 100 bp DNA Marker; lanes 2,3,4,5 and 6 positive mecAgene of MRSA strains (310 bp). Lane1: Control positive.
Sequencing of MecA Gene
Purification and standard forward sequencing of MecA gene were done by ABI Genetic Analyzer (Applied Bio systems). Five products selected to detect mecA gene sequences of PCR products in (Macrogens Inc, South Korea) at both directions with the same set of primers used for the PCR by Sanger dideoxy chain termination method (Figures 1 & 2).
Bioinformatics Analysis
The chromatogram sequences were visualized using Finch TV program version 1.4.0. The nucleotide sequences of the Mec A gene were searched for sequences similarity using online BLASTn 13. Highly similar sequences (accession number MH807092.1) was retrieved from NCBI Gen Bank and subjected to multiple sequence alignment using Bio Edit software version 7.2.514.
Results
A total of 100 eye swabs isolates were identified as S. aureus by conventional methods, including growth characteristics, colony morphology, and biochemical tests. The results revealed that S. aureus were identified in 100 (23.5%) patients 53 (12.4%) were females and 47 (11.1 %) were males. S. aureus was found highly resistant to methicillin MRSA 89 (89.0%) and less resistant to vancomycin VRSA 6 (6.0%). All S.aureus isolates were positive for 16SrRNA. All 89 methicillin-resistant S. aureus (MRSA) and 11 methicillin-sensitive S. aureus (MSSA) strains were tested for the presence of mecA 30/89 (33.7 %) were positive for mecA gene. Whereas the MSSA isolates were negative for the mecA gene. 5 samples (13, 17, 52, 58 and 140) were sent for characterization by sequencing of PCR products. The analysis of the mecA sequence revealed that there is no obvious mutation in the gene. BLAST search at the Gen Bank database with the mecA sequences for other species of S. aureus displayed that were clearly closely related to MH807092.1, from India respectively with a nucleotide sequence identity of 100 % as shown (Figure 3).
Figure 3: A. Electrophotogram of mecA gene,
B. Nucleotide sequence of mecA gene,
C. Alignment Amino acid sequence alignment of mec Agene,
D. Result of alignment of mecA amplicon with Staphylococcus aureus strain MecA-S1 MecA MH807092.1,
E. Result of Bio Edit program.MH807092, Staphylococcus aureus strain MecA-S1 MecA protein gene, partial acids.
Discussion
S. aureus is one of the most common causes of nosocomial infections and highly prevalent in Khartoum hospitals (Sudan). MRSA was highly (89 %) prevalent among populations of Staph. aureus isolated from conjunctivitis patients in different hospitals. Globally the presence of the mecA gene is considered the hallmark for identification of MRSA strains as found in Sudan by Ahmed et al., 2014, in Jordan by Alzubi et al., 2004, in Indian by Malathi et al., 2009 and in Saudi Arabia by Madani et al 200115,16,11,17. The findings in the present study showed low frequency of the mecA gene (33.7%); this may open the door to search for other intrinsic factors that may compete mecA gene in generating the increased resistance phenomenon in sudan with high prevalence of MRSA. The absence of mecA in MRSA strains has been reported by many authors worldwide Garica et al 2011and Ektik et al 20182, 18. This finding is consistent with the fact that penicillinase hyper producers do not show heteroresistance, The expected complement of genes that encode PBP may present (PBP1, PBP2, PBP3, and PBP4) in a highly conserved form compared with other sequences from other S.aureus isolates (data not shown), and we recorded no additional PBPs2. The S. aureus strains that are mecA-negative and exhibit oxacillin- or cefoxitin resistance did not harbor the mecC gene; therefore, they could carry other variations of the mecA gene that are not as well known or could present uncommon phenotypes such as borderline oxacillin resistance. Therefore, the whole-genome sequencing is always necessary to understand the mechanism of resistance 19.
Conclusion
In conclusion, Mec A gene -based PCR assay and sequencing are highly specific, sensitive and reliable method for identification of Methicillin-resistant Staphylococcus aureus (MRSA) and its differentiation from other genotypic closely related Staphylococcus species.
Data availability
The results of the nucleotide sequences of the Mec Agene were submitted in the GenBank database. Accession number: MH807092.1.
References
- Bamigboye BT, Olowe OA, Taiwo SS (2018) Phenotypic and molecular identification of vancomycin resistance in clinical Staphylococcus aureus isolates in Osogbo, Nigeria. European Journal of Microbiology and Immunology 8(1): 25-30.
- García Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, et al. (2011) Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. The Lancet infectious diseases 11(8): 595-603.
- Agha NFS (2020) Conjunctivitis among Rural and Urban School Children in Erbil Governorate/Iraq. Diyala Journal of Medicine 18(1): 58-69.
- Cavuoto K, Zutshib D, Karp C, Miller D, Feuer W (2008) Update on bacterial conjunctivitis in South Florida. Ophthalmology 115(1): 51-56.
- Rasheed NA, Hussein NR (2020) Methicillin-resistant Staphylococcus aureus carriage rate and molecular characterization of the staphylococcal cassette chromosome mec among Syrian refugees in Iraq. International Journal of Infectious Diseases 91: 218-222.
- Abulreesh HH, Organji SR, Osman GE, Elbanna K, Almalki M, et al. (2017) Prevalence of antibiotic resistance and virulence factors encoding genes in clinical Staphylococcus aureus isolates in Saudi Arabia. Clinical Epidemiology and Global Health 5(4): 196-202.
- Velasco V, Vergara JL, Bonilla AM, Munoz J (2018) Prevalence and characterization of Staphylococcus aureus strains in the pork chain supply in Chile. Foodborne pathogens and disease 15(5): 262-268.
- Haaber J, Penadés JR, Ingmer H (2017) Transfer of antibiotic resistance in Staphylococcus aureus. Trends in microbiology 25(11): 893-905.
- San Sit P, The CSJ, Idris N, Sam IC, Omar SFS, et al. (2017) Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) infection and the molecular characteristics of MRSA bacteraemia over a two-year period in a tertiary teaching hospital in Malaysia. BMC infectious diseases 17(1): 274.
- (2010) CLSI: Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twentieth informational supplement ed. CLSI document. 2010; M100-S20 Wayne, PA, USA.
- Malathi J, Sowmiya M, Margarita S, Madhavan HN, Therese KL (2009) Application of PCR based--RFLP for species identification of ocular isolates of methicillin resistant staphylococci (MRS). Indian Journal of Medical Research 130(1): 78.
- Saha B, Singh AK, Ghosh A, Bal M (2008) Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). Journal of medical microbiology 57(1): 72-79.
- Seth Smith, HM Rosser, SJ Basran, A Travis ER, Dabbs ER et al. (2002) Cloning, sequencing, and characterization of the hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine degradation gene cluster from Rhodococcus rhodochrous. Applied and Environmental Microbiology 68(10) 4764-4771.
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic acids symposium seriesLondon]: Information Retrieval Ltd c1979-c2000 41(41).
- Abdelgadeir LM (2015) Van B Positive Vancomycin-resistant Staphylococcus aureus among clinical isolates in Shendi City, Northern Sudan. IOSR J Dental Med Sci 14: 87-91.
- AlZubi E, Bdour S, Shehabi AA (2004) Antibiotic resistance patterns of mecA-positive Staphylococcus aureus isolates from clinical specimens and nasal carriage. Microbial Drug Resistance 10(4): 321-324.

 Review Article
Review Article