Abstract
Many methodologies, as well as information systems, have been developed to support medical decision making in terms of disease diagnosis. The majority of them are based on keyword searches, while others on artificial intelligence, ontologies, etc. However, there is no single approach that can successfully diagnose diseases because most of them rely on either the statistical appearance of keywords describing symptoms, and many have similar symptoms. In contrast, the more advanced ones dealing with artificial intelligence approaches and ontologies lead to poor results. This paper proposes a system based on the approach given by the medical practice itself and is called Differential Diagnosis (DD). More to the point, doctors’ logic concerns data retrieval from knowledge recorded in their memory, combining thus more descriptive expressions that describe symptoms critical for concluding the correct disease. One doctor could exclude diseases or converge on them depending on the combined data assessment from the clinical examination and the history, which is then confirmed by some additional examinations. The proposed system approaches the diagnosis with the logic of the doctor’s approach due to the DD. It relies on its back-end core where Natural Language Processing (NLP) of the medical bibliography is utilized. Furthermore, common areas, with medical protocols and findings on Venn diagrams leading to a successful diagnosis, are extracted.
Keywords: Differential Diagnosis; Natural Language Processing; Information Retrieval; Venn Diagram
Abbreviations: DD: Differential Diagnosis; NLP: Natural Language Processing; ICT: Information and Communication Technologies; EMRs: Electronic Medical Records
Mini Review
DD constitutes the process of disease detection, among several diseases, which presents similar clinical findings and symptoms. Doctors apply diagnostic procedures either to diagnose the disease or to approach the potential disease. It is a systematic diagnostic method for diagnosing the disease in a patient. It may not be clear to make a safe diagnosis due to many diseases that show small differences in subtle symptoms. Different methodological approaches can be applied to DD, based on a process of elimination of the least probable diseases according to sets of criteria according to the knowledge of the doctor. Medical logic follows a process of information retrieval that reduces the chances of the parameters of several candidate diseases to negligible levels based on symptoms, patient history (chief complaint, history of present illness, past medical history, past surgical history, social history, medications, and allergies, etc.), laboratory tests, etc. and, of course, medical knowledge to approach the most likely disease [1-3].
Many Information and Communication Technologies (ICT) tools promise to help and support a medical decision in diagnosing diseases. In most of these tools, the doctor should enter the symptoms that find in the clinical examination of a patient. Usually, these symptoms can be entered into the system as one word, i.e., fever, headache, etc. Many diseases have similar symptoms, and as a result, many diseases appear on the doctor’s screen, usually in terms of information retrieval in the system’s databases. For example, it is difficult for an information system to distinguish low-temperature fever from high-temperature fever. The low-temperature fever can lead to a cold disease, while a high-temperature fever can lead to the flu. Many of them also indicate the possibility of developing a disease, and in following, the doctor identifies many diseases with equal possibility, i.e., influenza 80%, cold 80%, etc. which means that eight out of ten symptoms, which doctor entered, were found in these diseases. The basic logic of these tools is implemented by matching the keywords (symptoms) that the doctor places in the system and obviously, the output is the display in the form of percentages of the number of criteria that match a disease on all the symptoms set by the doctor [4,5].
Other approaches regarding differential diagnosis are based on artificial intelligence. These systems for medical decision support are more likely to be found in clinical laboratories and educational settings in terms of clinical surveillance or areas with a huge number of data, like an intensive care setting. More specifically, if a corresponding appropriate rule is taken into account, then intelligent programs can indeed offer significant benefits. These systems can be classified into two categories: rule-based expert systems and those based on probabilistic graphical models, often called probabilistic expert systems or normative systems [6]. The evolution of Natural Language Processing (NLP), along with big data applications, the emergence of compelling computers, the cloud computing infrastructure, and the improvement of NLP algorithms create new possibilities in search of content in digital texts. Nowadays, ICT solutions can analyze more language-based data than humans themselves. Considering the huge amount of unstructured data generated every day, automation will be critical to analyze text efficiently thoroughly.
Similarly, deep learning is now widely used for modelling human language while, on the other hand, the need for semantic understanding is not necessarily present in these machine learning approaches. NLP is playing an essential role as it helps in resolving language ambiguity and adds useful numeric structure to the data for many downstream applications, such as speech recognition or text analytics. In the scientific area of medicine, texts found in the bibliography, in scientific publications, in information leaflets, in medical records, etc. can be considered as big data information, where doctors must have the latest information and be continuously updated [7-11]. The increasing adoption, the evolving and different formats of Electronic Medical Records (EΜRs), as well as the great interest and need for using these data to support doctors in clinical work, have made clear that the data retrieval of narrative text contained in the medical records and bibliography is unavoidable.
On the other hand, the medical bibliography and publications are considered important sources of information and should be approached from many aspects of information technologies; one example is the NLP, because the medical texts contain complex terminology that complicates the classical techniques applied to other cases of text and data extraction [11]. However, all previous approaches have been based on the logic of storing and retrieving medical data, essentially providing a statistical response on the set of symptoms for diseases that match the keywords selected by the doctor from a default list. Moreover, the result of each tool is simply a group of potential diseases corresponding to a portion of selected keywords. The result of large lists given diverse diseases with equal probability reminds the doctor of existing diseases without supporting the medical decision. This is also why most of the previous approaches and tools have almost no penetration into the medical practice and no impact on doctors.
The doctors will benefit from our proposed support system as they usually prefer to describe the patient’s clinical findings and history. This enables doctors not to change their habits and daily life as many data entry fields can be answered; for example, symptoms can be placed from an existing list of symptoms, in following images of internal organs, etc. Also, free text can be utilized, and natural text and the data retrieval process will be based on NLP techniques. The back-end modules combine the data entered and inserted by the doctor through a user-friendly interface, then retrieve the probable diseases by the clinical findings as well as patient’s history with the use of DD methodologies for selecting the most likely diseases, and finally suggest to doctors selected laboratory assessments, imaging tests, and histopathological tests. At each step, the novel approach of incorporating Venn diagrams focuses on the most probable diseases, while grading the diseases with lower probabilities. The system suggests selected additional tests that safely confirm the assessment of the diseases. Finally, this medical decision support system lists treatment and medication, which have been identified in the medical bibliography and publications.
Proposed System
Successful Diagnosis Parameters
The approach based on DD for the medical side approach and an advanced information system that will support the medical decision and prescription is proposed. The corresponding approach will combine the parameters (symptoms). Each time the doctor is called to consider with in-depth search and correlation of knowledge to lead to diagnosis and appropriate treatment. DD, as the diagnostic process through which diseases with similar symptoms are ruled out, aims to help at the predominant level of diagnosis. The process involves monitoring and discovering factors and symptoms, then gradually calculating the most likely diseases; in the following ruling out a few possible diagnoses, one after the other until, in the end, there is only one specific diagnosis that explains all the signs, symptoms of the patient. Therefore, in medicine, the successful diagnosis leads to early treatment and requires the acquisition and capture of the image resulting from the clinical examination, as well as the successful recording of the medical history and the recording of the data, and finally, the execution- preferably- directed and targeted (based on history and clinical examination) laboratory tests.
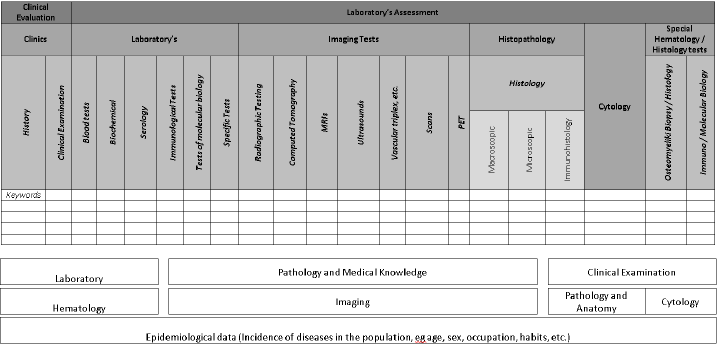
The proposed system will record medical history, being the potential to both correlate and investigate/clarify and save classical medical knowledge, which is the knowledge thesaurus of the system. Furthermore, from this thesaurus, correlated data and information/knowledge according to data from the history (for this disease) will be drawn and crosschecked. Also, findings of physical examination and laboratory data such as blood, biochemical, serological tests, molecular biology tests, and other laboratory tests, imaging tests (radiography, CT, MRI, scans, PET, etc.), histological and serological tests, as well as specific blood tests such as biopsy, immunophenotype, genetic testing, etc. will also be extracted. To further achieve rational diagnosis and treatment, the performance of the perfect history and clinical examination is expected to reach the probability of 60% in terms of correct diagnosis and subsequent treatment. Based on the execution of these laboratory tests, the possibility of achieving the proper diagnosis and next treatment is increased by 20% (total 80% successful diagnosis), thus remaining an unspecified probability with a percentage equal to 20%.
The information system mainly aims at the improvement of achieving successful diagnosis and treatment by improving speed and eliminating the uncertainty of this unspecified rate through correlation of the parameters and deep search as well as correlation with clinical and laboratory data. The history of each patient, their clinical examination, and all laboratory data consist of new parameters at each specific time period and characterize the current pathology of our patients. Based on the above, the medical brain data is stimulated, so after vertical, horizontal, and crosscorrelations, the doctor will put the information system into deep search and correlation knowledge. As a result, the final correlations in those three levels and searching for the stored knowledge can lead to diagnosis and appropriate treatment (Figure 1). The doctors enter text in the patient’s history in terms of electronic forms. Therefore, the natural text analysis, along with the data retrieval process, is based on NLP techniques, which will be analyzed in the following subsection.
Back-end System using NLP in Medical Bibliography and Publications
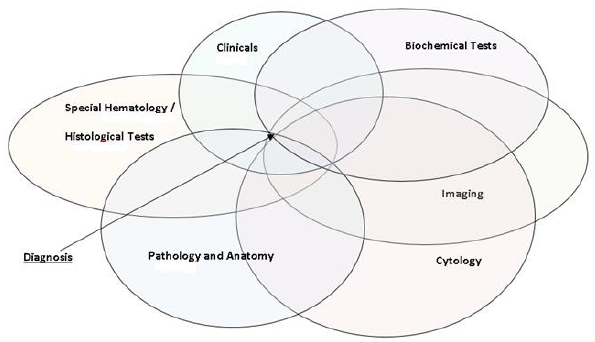
For assessing the medical data and the parameters entered into the system, all the recorded data are integrated to form a Venn diagram. The information retrieval from the medical literature for the Venn diagram construction will be implemented with Natural Language Processing (NLP) methodologies to extract medical information for the back-end module of the system. Each parameter is associated with each column vertically, horizontally, and crosswise with all positive (outside normal limits) in each column parameters naming the content of each cycle Venn diagram. The deepest knowledge of thesaurus is to be associated with pathological conditions or diseases that reflect the set of pathological parameters as listed in the history, the clinical examination, and the laboratory assessment.
The first diagnosis or set of diagnoses will be the one that has a common area. The ideal diagnosis is related to the Venn diagram. In contrast, the second diagnosis will have a common area suitable for five cycles, etc. until only one parameter cycle is left, representing the latest possible diagnosis item. So, the number of diagram cycles (with parameters in each cycle) depicts the content and the range of possible diagnoses that can be considered in order. In this way, possible diagnoses can be prioritized in terms of the cycles of the diagram that have a common area. In such cases (based on the recorded parameters from the overall medical assessment of the patient), fewer than six cycles of Venn diagrams, involved in the common area, may be inferred diagnoses series with a specific area in the diagram. Consequently, the hierarchy of diagnoses may be supported by the evaluation and correlation of incidence, frequency diseases, and diagnoses in the study population, according to published epidemiological parameters such as geographical localization, age, sex, occupation, etc. So, the final diagnosis can reflect the reality of the statistics.
Assessment and Correlation of Back-end Information
For the assessment of data from these columns, all the above parameters are recorded if their values exceed the normal limits of each column. What is more, the data is integrated to form a Venn diagram. Each parameter is associated with each column vertically, horizontally, and crosswise with all positive (outside normal limits) in each column parameters naming the content of each cycle Venn diagram. The deepest knowledge of the thesaurus is associated with pathological conditions or diseases that reflect the set of pathological parameters as listed in the history, the clinical examination, and the laboratory assessment.
Stimulus (pathological parameters: history, physical examination, laboratory evaluations). Deep Search (Deep correlation (vertical, horizontal, and cross combination) of the parameters and junction thesauri) (Figures 2 & 3).
System Modules
To visualize the parameters on a Venn diagram, an electronic medical record should be created where the data will be reflected in the table of the overall medical evaluation. The electronic medical records should be composed of fields that describe the detailed “history”, the “clinical examination”, the “laboratory”, the “imaging studies”, the “pathology”, the “cytology as well as the “special hematology/histology.” From this electronic medical record, all the necessary pathological parameters will be derived to implement the differential diagnosis according to the above methodology.
The proposed system consists of two main parts:
1) The system of input records and
2) The system of information processing.
The first part will be carried out with access from web-based technologies available on both mobile devices- which are the main focus of the clinical trial and the laptops and desktops and aimed at primary healthcare staff and primary healthcare units. The access method ensures direct access of users from any device, compatibility with all computer systems and easy use by web browsers, the ability to use the system independently of the location, etc.
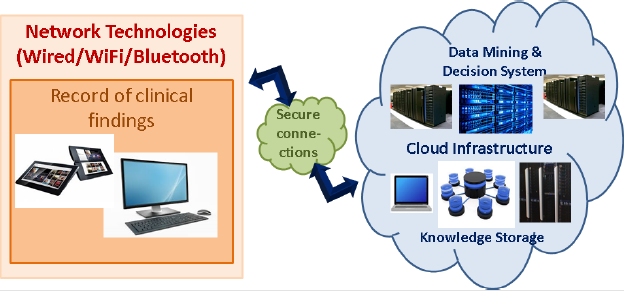
The front-end part, which will be provided to the doctor, will be able to use the three-dimensional (3-D) graphical environment to enter the findings of the clinical examination, in addition to type in the observations. Also, should the doctor deem it necessary, a form of focused and free recording of history will be completed. Using network technologies (wired and/or wireless), the recorded information (image, keywords, free text) will be sent to the back-end sub-system of the medical decision-support information system. It is a distributed large volume data management information system based on cloud computing technologies, with all the known literature and medical science references in appropriate structures. The search will be utilized with the use of algorithmic methodologies for retrieving large volumes of data (Big Data Text Mining). The structures will preserve a treasure trove of the clinical picture, the symptoms, the diagnoses, the appropriate treatment, the required laboratory, the imaging, the examinations required, the treatment reactions, and the precautions of any disease.
It is pointed out that the information regarding the medical decision will be distributed in terms of geographical medicine and seasonality, to speed up the retrieval and extraction of the results that support the medical decision and take place in real-time, facilitating the work of doctors. The back-end part of the system will have its interfaces that help researchers, support doctors, healthcare staff, and technical staff to be able to manage the system from all angles, thus evolving the system into a decision system based on machine learning. Furthermore, it will be enhanced with additional functions that can incorporate possibilities of correlation and investigation/clarification of the already stored classical medical knowledge with findings of laboratory and imaging that will follow the medical decision and once fed back into the system. It will help to learn and increase its success in future diagnoses (Figure 4).
Conclusion and Future Work
The proposed system will provide better healthcare services, improve patient’s conditions, provide faster diagnosis, reduce delays and potential mortality, reduce hospitalization costs by saving diagnostic time and hospitalization time by diagnostic delays, reduce system costs by proposals for focused laboratory and imaging confirmation tests, reduction of consumption of reagents, drugs and general medical equipment, optimal use of pharmaceutical material, etc.
The technological and scientific objectives for the next steps of the proposed system are:
a) The development of a clinically friendly mobile system for capturing clinical images and history via wireless networking,
b) The extraction of medical information, the discovery of knowledge and support of medical decision from a large amount of data of the medical literature, for the automated differential diagnosis of cases,
c) The creation of computer models for the most efficient support of the medical diagnosis,
d) The creation of support of laboratory and imaging examinations confirmation of the diagnosis,
e) The interconnection with existing hospital medicine management systems,
f) The security of the information system and its compliance with the requirements of European Regulation 2016/679 (General Regulation on Data Protection - GDPR).
References
- Henderson MC, Tierney LM, Smetana GW (2012) The Patient History: Evidence-Based Approach to Differential Diagnosis. McGraw Hill, New York, NY, USA.
- Siegenthaler W (2007) Differential diagnosis in internal medicine: from symptom to diagnosis. pp. 1140.
- Strain S, Franklin S (2011) Modeling Medical Diagnosis Using a Comprehensive Cognitive Architecture. Journal of Healthcare Engineering 2(2): 241-258.
- Alves R, Piñol M, Vilaplana J, Teixidó I, Cruz J, et al. (2016) Computer-assisted initial diagnosis of rare diseases. PeeJ 4: e2211.
- Schiza E, Panos G, David C, Petkov N, Schizas C (2015) Integrated Electronic Health Record Database Management System: A Proposal. Stud Health Technol Inform 213: 187-190.
- Adewole KS, Hambali M, Jimoh MK (2015) Rule-based Expert System for Disease Diagnosis. Proceedings of the International Conference on Science, Technology, Education, Arts, Management and Social Sciences (iSTEAMS Research Nexus).
- Spasic I, Ananiadou S, Mc Naught J, Kumar A (2005) Text mining and ontologies in biomedicine: making sense of raw text. Briefings in Bioinformatics 6(3): 239-251.
- Ferré A, Ba M, Bossye R (2019) Improving the CONTES method for normalizing biomedical text entities with concepts from an ontology with (almost) no training data. ISSN 2234-0742. Genomics Inform 17(2): e20.
- Hai Long T, Nguyen NTH, Makoto M, Ananiadou S (2018) Investigating Domain-Specific Information for Neural Coreference Resolution on Biomedical Texts. Proceedings of the BioNLP 2018 workshop, pp. 183-188.
- Indu N, Aswathy PR (2019) Natural Language Processing in Medicine: A Review. International Journal of Engineering Research & Technology (IJERT) 7(5): 1-4.
- Dhole G, Uke N (2014) NLP based retrieval of medical information for diagnosis of human diseases. IJRET: International Journal of Research in Engineering and Technology 3(10): 243-248.

 Mini Review
Mini Review