Abstract
Single nucleotide polymorphism (SNP) analysis has emerged as the most relevant method in DNA profiling. Forensically relevant SNP markers have been employed in the present research to unveil variations in ethnic individuals employing a set of 200 PCR amplicons. Sequencing results revealed polymorphism at single nucleotide level in different samples when compared to the already reported ones. Multiple sequence alignment of various samples from ancestry informative markers (rs713367 and rs34940277) exhibited in alteration of nucleotide A to G and GA to AG, respectively. Variation of G was found with A/C in case of phenotypic informative marker (rs199920775). Data for identity informative primers rs1542931and rs1988436 revealed substitution of nucleotide T to C and A, respectively. In case of lineage informative marker rs3908, deletion was observed for nucleotide G. All variations found were synonymous with respect to coding consequences, which might directly impact on function of gene through diverse cellular mechanisms. The data collected is an initiative to facilitate forensic DNA investigation and to cover gaps in DNA profiling in Pakistan if linked with latest biometric computerized National Identity Card system.
Keywords: Single Nucleotide Polymorphism; Sequencing; Variation Analysis; Forensics; Pothwari
Introduction
Single Nucleotide Polymorphism (SNPs) being simplest form of variation promises the assistance for forensic DNA analyses because of an excess of potential markers, its automation, and reasonable reduction in required fragment length[1]. SNPs have vital role in causing diversity among individuals, phenotypic traits such as hair texture/ color, skin tone, eye color, nose/ear shape etc., difference in drug response among individuals, diseases, evolution etc. in nonsynonymous/ synonymous changes, mRNA stability, gene/ protein expression etc[2]. SNP analysis proves to be more beneficent as compared to STR typing, in dealing with highly degraded biological materials, in some situations including mass disasters, missing persons and unidentified human remains where the DNA may be substantially fragmented, mtDNA[3]Y chromosome study for lineage information purpose, biographical ancestry analysis [4], power to identify phenotypic characteristics [5].
Standardization and inter laboratory validation assays will be key for the use of SNPs in the forensic field[6]. As SNPs have relatively low mutation rates so these meant to be more authentic genetic markers for providing investigative information in some exceptional cases [7]. As per forensic application, SNPs have categorized into 5 different types. These include identity-informative SNPs (IISNPs) [8-11] for recognition purposes, lineage-informative SNPs (LISNPs) [12]for inferring paternity (especially useful in kinship analysis and paternity testing), ancestry-informative SNPs (AISNPs) [9,13] for ancestry characterization, and for identification of phenotypic attributes, phenotypic-informative SNPs (PISNPs) [14-17].
Crimes that can be solved through forensics are common in Pakistan. As per current situation and especially in last few years, Pakistan has been engaged in fighting against many hazarded factors such as terrorists attack, man-made as well as natural disasters, military conflicts, crimes etc[18]. An inclusive DNA database lacks in Pakistan which must be established to match samples from crime scenes against already existing evidence. Pakistani government is attempting to develop DNA database of all its citizens at national level keeping in account the stronger desires of Pakistani citizens for DNA profiling. The data in DNA databases can be linked with latest biometric computerized National Identity Card (NIC) system that can facilitate not only in searching out criminals and bombers but to identify victims of mishaps. In Lahore, world’s second largest forensic laboratory (Punjab Forensic Science Agency) with excellent facilities for forensic examination in Pakistan established by the Punjab government to counter terrorism, is trying to collaborate and associate with world’s eminent forensic institutes for strengthening and growth of laboratory [19].
Pakistan is a country with diverse ethnic groups. Therefore, exploitation of genetic diversity through forensic DNA markers may be a significant attempt to generate the DNA profiles of different populations across Pakistan for record and investigation of case using DNA markers. The major aim of the present study was to amplify the forensically relevant loci from Pothwari population with different types of SNP markers. The ultimate purpose was to perform the sequencing and analysis of amplified amplicon with different bioinformatics tools to infer the forensically relevant SNP variations existing between individuals in Pothwar region. Through present study contribution has been made in adding up useful information related to polymorphism and variations. This piece of knowledge can benefit researchers in their special training in forensics, communication and collaboration with different inter and intra country forensic research institutes.
Materials and Methods
Region of Present Study
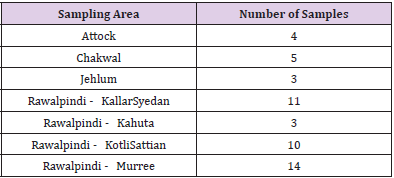
Ethnic individuals residing in Pothwar region were selected for present research. Pothwar/Panjistan region located in North- Eastern Pakistan, covers the Northern side of Punjab. The Western areas of Azad Kashmir and the Southern parts of Khyber Pakhtunkhwa are at its borders. The Pothohar Plateau includes the four districts namely Jhelum, Chakwal, Rawalpindi and Attock[20]. For identification of individuals from Pothwari population parameters were set as criteria such as ethnicity, birthplace (of individual and forefather’s), and first language. All data and statistics were documented in the “Consent Form”.
Sample Preparation
5ml of blood was drawn using BD syringes (5ml) in EDTA tubes by a trained professional from 50 unrelated healthy male individuals and stored at 4˚C in the laboratory before being processed for extraction of genomic DNA. Sampling detail has been summarized in Table 1.
Extraction of Genomic DNA and PCR
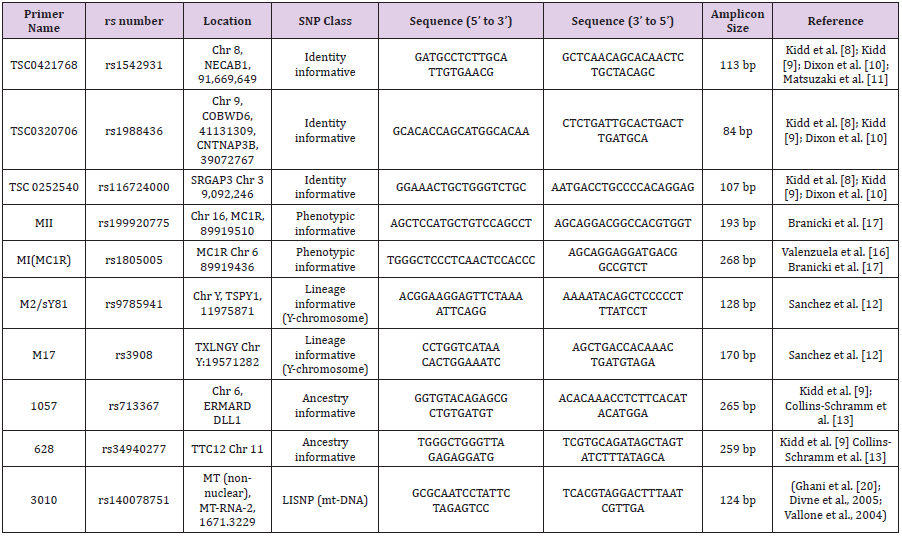
DNA extracted from blood using PureLink™ Genomic DNA Kits (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA). Concentration and purity of extracted DNA was checked through NanoDropTM 1000 spectrophotometer. Primers were selected from five distinct categories of SNPs, list of which with their complete detail is given in Table 2. For this purpose, literature was reviewed and online databases such as STRbase[21], SNPCheck (Lai and Love, 2012), UCSC In Silico PCR [22], dbSNP[23], were fetched. A total volume of 50 μLreaction was prepared for PCR. Thermal cycling was performed at conditions of denaturation at 95°C for 5 min followed by 37 cycles of denaturing at 94°C for 40 s, annealing set in accordance to primer’s Tm values for 1 min, and extension at 68°C for 1 min. Samples were held at 4°C. Products of DNA and PCR were examined on 1% and 1.5% agarose gel, respectively stained with ethidium bromide.
Sequencingof Amplified Products
A set of 200 PCR amplified samples was prepared. This dataset was prepared using ten forensically relevant SNPs markers from five distinct categories against 50 samples. Sanger sequencing of these PCR amplicons was done through a commercial company (Macrogen Korea).
Identificationof SNPs Variation and their Consequences
Outputs of sequencing were subjected for various analysis steps to study SNP variation. These include trimming, editing, alignment, mutational studies, identity and similarity etc. employing various tools and software such as ClustalW, Molecular Evolutionary Genetics Analysis MEGA Version 7.0 [24]BioEdit[25], DNAsp[26]. Consequences of variants were analyzed using variant effect predictor tool [27].
Results
Forensically Relevant SNP Markers Amplified from Selected Samples
Extracted DNA products with satisfied values of Purity (~ 1.80) and concentration (260/280 ratio) were used further for amplification step (Figure 1). The amplicon length was different for different SNP primers. Amplification results justified these threshold values as each product length was in accordance to the actual primer amplicon size which was analyzed with the help of ladder. A maximum size product of 268bp for rs1805005 was obtained in contrast with rs116724000 where it was only 107bp. A PCR product of 268bp, 265bp, 259bp, 193 bp, 170bp, 128bp, 124bp, 113bp, 107bp was obtained for rs1805005, rs713367, (rs34940277), (rs3908), rs199920775, rs9785941, rs1988436 and rs116724000, respectively. For rs1988436, rs1988436 and rs140078751, same size amplicons were obtained i.e. 124bp. Sequencing files were received in pdf, notepad, FASTA, trace. abi and phd.1 formats.
Figure 1: Visualization of PCR Amplicons under UV gel documentation system: Electropherogram of ethidium bromide stained 1.5 % agarose gel a) rs34940277, b) RS1805005, c) rs116724000 d) rs3908 e) rs140078751.
The raw sequences obtained were searched against the program Blastn at NCBI. The sequences hitting the target SNP marker locus were selected for further analysis. The target sequences were aligned and analyzed using Bio-Edit and MEGA 7 (Molecular Evolutionary Genetic Analysis Version 7) software. The sequences were trimmed to a size according to the amplicon length. The first 20 and last 20 bases at the 3’end were whittled downed. The trimmed sequences were aligned using MEGA 7 software.
Sequence Analysis Reveal SNP Variations in Samples
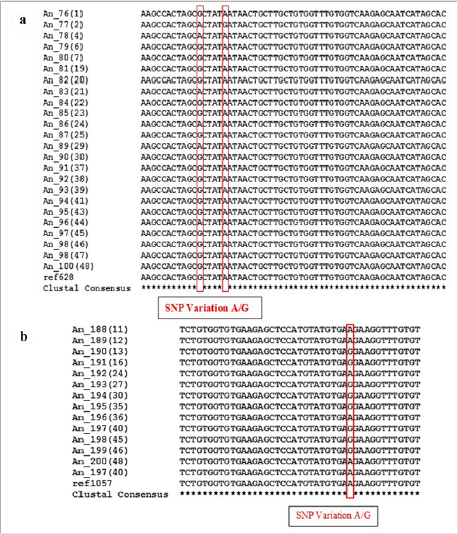
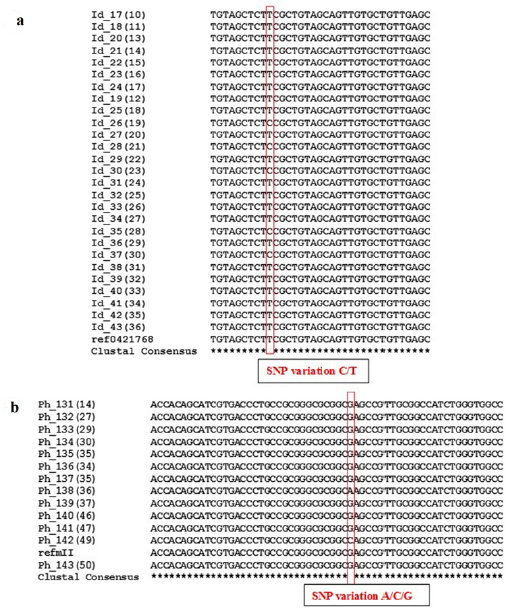
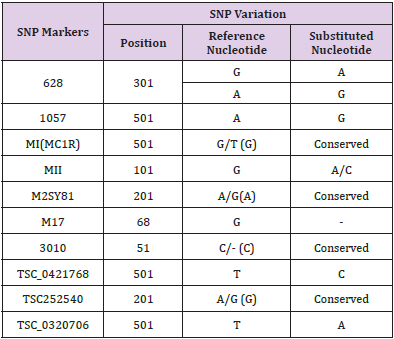
In order to identify the SNPs amongst trimmed samples, sequences were directed to multiple sequence alignment using ClustalW Program in MEGA7. The alignments revealed conservation among all the sequences but variations at few points were also observed. Table 3 shows detail of SNP variations which have been observed in results obtained. This result shows existence of SNPs in population for rs1988436, rs713367, rs199920775, rs34940277, rs3908, and rs1542931. Results for ancestry informative primers have been shown in Figure 2 which shows polymorphism of G to A in six samples in comparison to reference for rs34940277 and for rs713367 in 8 samples from a total of 14, SNP (A to G change) has been observed. Alignment for identity & phenotypic primers i.e. rs1542931 & rs199920775, respectively has been illustrated in Figure 3. rs1988436 shows single nucleotide polymorphism of T to A in eight samples out of a total 29 (Figure 4) with respect to reference sequence. Results for remaining categories has been provided in supplementary material.
Figure 2: Multiple Sequence Alignment using MEGA7 showing SNP variations in Ancestry Informative Primers highlighted with red rectangle: a) Polymorphism for rs34940277 found at two positions with respect to reference sequence i.e. G substitution with A and substitution of A with G in case of sample An_77(2). b) Results for Primer rs713367 showing variation of A in reference sequence with G in various samples under study. Conserved Regions indicated by asterisk (*).
Figure 3: MEGA7 Alignment showing SNP variations highlighted: a) Identity informative primer rs1542931 highlighting SNP variation of T in reference sequence with C in various samples b) Phenotypic Informative marker rs199920775 showing substitution of G in reference sequence with A and C in samples Ph_138(36) and Ph_142(49), respectively.
Figure 4: SNP variations analysis: a) Results for identity informative primer rs1542931 using MEGA7 highlighting SNP variation of T in reference sequence with C in various samples b) Results for Phenotypic Informative marker rs199920775 showing substitution of G in reference sequence with A and C in samples Ph_138(36) and Ph_142(49), respectively.
SNP variation indicates existence of diversity among different individuals from same population. In case of markers rs1805005, rs9785941, rs140078751, rs116724000, no SNP variation was observed as all nucleotide sequences of samples were completely aligned with reference sequence i.e. 100% conservation. Absence of SNP variation shows that the sequence is conserved and there is no variation among different individuals from same population for these primers. Detail for each primer category is given in Table A provided as a supplementary material.
Variant Effect Predictor Annotation
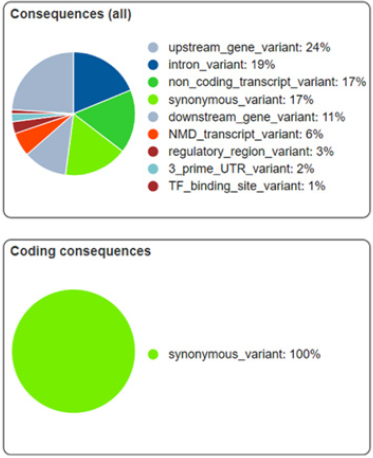
Variation has been found against six markers, consequences of which have been illustrated in Figure 5. This demonstrates each category of variants with distinctive colors. Coding consequences for these variants have been observed as 100% synonymous with no variant lying in non-synonymous category which has also been cross confirmed using DNAsp. This implies that the SNP variations among sequences are not present in coding region therefore does not affect the gene function and its expression however regulation might have relation with it which brings diversity. Minimum percentage is of transcription factor binding site variant i.e. 1% and upstream gene variants are maximum in number i.e. 24%.
Figure 5: Summary for Consequences of variants. Upper panel showing percentage wise types of effects and lower panel is showing percentage variants as synonymous or non-synonymous.
Discussion
Present results obtained from a dataset of 200 samples using forensically relevant SNP markers show that SNP variations are present in individuals of Pothwari population. These loci were selected to identify sequence pattern and to check whether the SNP exist in pothwari individuals. Variations have been observed against 6 primers rs1988436, rs713367, rs199920775, rs34940277, rs3908, and rs1542931 and all these were synonymous, which implies that the SNP variations among sequences are in noncoding region which can have direct impact on function of gene through diverse cellular mechanisms. 100% conservation was also observed against rs9785941, rs140078751, rs1805005, and rs116724000 which shows that the sequence is conserved and there is no SNP variation among different individuals for these markers. DNA analysis provides basic foundation for contemporary forensic research.
Work on SNPs as presented in the current study is tremendously useful and has also remained the focus of different researchers who published their research efforts. Phillips in 2004 picked and worked on autosomal SNPs, mtDNA coding region SNPs and Y-chromosome SNPs and almost 10 individuals with 10 additional attributes were identified with SNP analysis alone, when SNP genotypes were used to supplement partial STR profiles[28]. Work has been done in 2008 to investigate the genetics of the human Mediterranean populations and migration rate studies, using SNPs located on the sex chromosomes[22]. In 2014, four new polymorphic positions 11,741, 11,756, 11,878, and 12,133 in mtDNA were detected through multi-locus association between 25 SNPs of X-chromosome for Ibiza and Cosenza populations which proved to be a source for human identification purpose in Iraq [29]. In 2015 Santos with his team developed Pacifiplex which is a sensitive multiplex assay, comprising 29 ancestry-informative marker SNPs to complement the 34-plex test that distinguished Africans, Europeans, East Asians and Oceanians in a combined set [30].
Wang and Moult during their research in 2001 have analyzed the effect of a set of disease-causing missense mutations emerging from SNPs, and a newly determined SNPs set from the common population and were successful in developing a model for assigning a mechanism of action of each mutation at the protein level [31]. Information collected in the current study is useful and can definitely facilitate analysis from different aspects such as in accordance with disease relevance, will make possible forensic DNA testing, and can be used as a part of record for investigation in case of any mishap or disaster. It will overcome flaws in DNA profiling in Pakistan and relevant research work as per need of time. SNP analysis being key part of the investigation and experimental analysis is continuously solving the complex queries. The data obtained can benefit researchers, in situations of war & terror, mass disasters, mishaps, to answer complex situations and will facilitate forensic DNA investigation and cover gaps in DNA profiling in Pakistan.
Acknowledgement
We are thankful to Genome Editing & Sequencing Lab, National Center for Bioinformatics, Quaid i Azam University Islamabad, for providing us a working platform. We are grateful to all the volunteers for providing us samples. Their contribution made this research come to a conclusion.
References
- Yang Y, Xie B, Yan J (2014) Application of next-generation sequencing technology in forensic science. Genomics Proteomics Bioinformatics 12(5): 190-197.
- Shastry BS (2009) SNPs: Impact on Gene Function and Phenotype. In: Komar A. (eds) Single Nucleotide Polymorphisms. Methods in Molecular Biol 578: 3-22.
- Zambelli F, Vancampenhout K, Daneels D, Brown D, Mertens J, et al. (2017) Accurate and comprehensive analysis of single nucleotide variants and large deletions of the human mitochondrial genome in DNA and single cells, Eur J Hum Genet 25(11): 1229-1236.
- Kayser M (2017) Forensic use of Y-chromosome DNA: a general overview. Hum Gene 136(5): 621-635.
- Parsons TJ, Coble MD (2001) Increasing the forensic discrimination of mitochondrial DNA testing through analysis of the entire mitochondrial DNA genome. Croat Med J 42(3): 304-309.
- Mortera J, Dawid AP, Lauritzen SL (2003) Probabilistic expert systems for DNA mixture profiling, Theor Popul Biol 63(3): 191-205.
- Sobrino B, Bríon M, Carracedo A (2005) SNPs in forensic genetics: a review on SNP typing methodologies. Forensic Sci Int 154(2-3): 181-194.
- Kidd KK, Soundararajan U, Rajeevan H, Pakstis AJ, Moore KN, et al. (2018) The redesigned Forensic Research/Reference on Genetics-knowledge base, FROG-kb. Forensic Sci Int Gene 33: 33-37.
- Kidd KK, Pakstis AJ, Speed WC, Grigorenko EL, Kajuna SL, et al. (2006) Developing a SNP panel for forensic identification of individuals. Forensic Sci Int 164(1): 20-32.
- Dixon LA, Murray CM, Archer EJ, Dobbins AE, Koumi P, et al. (2005) Validation of a 21-locus autosomal SNP multiplex for forensic identification purposes. Forensic Sci Int 154(1): 62-77.
- Matsuzaki H, Dong S, Loi H, Di X, Liu G, et al. (2004) Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods 1(2): 109-11.
- Sanchez JJ, Børsting C, Hallenberg C, Buchard A, Hernandez A, et al. (2003) Multiplex PCR and minisequencing of SNPs--a model with 35 Y chromosome SNPs. Forensic Sci Int, 137(1): 74-84.
- Collins Schramm HE, Chima B, Morii T, Wah K, Figueroa Y, et al. (2004). Mexican American ancestry-informative markers: examination of population structure and marker characteristics in European Americans, Mexican Americans, Amerindians and Asians. Human Gene 114(3): 263-271.
- Speed D, Cai N, Johnson MR, Nejentsev S, Balding DJ, et al. (2017) Reevaluation of SNP heritability in complex human traits. Nat Genet 49(7): 986-992.
- Mehta B, Daniel R, Phillips C, McNevin D (2017) Forensically relevant SNaPshot® assays for human DNA SNP analysis: a review. Int J Leg Med 131(1): 21-37.
- Valenzuela RK, Henderson MS, Walsh MH, Garrison NA, Kelch JT, et al. (2010) Predicting phenotype from genotype: normal pigmentation. J Forensic Sci 55(2): 315-322.
- Branicki W, Brudnik U, Kupiec T, Wolañska Nowak P, Wojas-Pelc A (2007) Determination of phenotype associated SNPs in the MC1R gene. J Forensic Sci 52(2): 349-354.
- Kayani SA (2011) Global War on Terror: The Cost Pakistan is paying. Margalla Papers pp. 1-16.
- Zar MS, Shahid AA, Shahzad MS (2013) An Overview of Crimes, Terrorism and DNA Forensics in Pakistan. J Forensic Res 4(4): 1-2.
- Ghani MW, Arshad M, Shabbir A, Shakoor A, Mehmood N, et al. (2013) Investigation of potential water harvesting sites at potohar using modeling approach. Pak J Agri Sci 50(4): 723-729.
- Ruitberg CM, Reeder DJ, Butler JM (2001) STRBase: a short tandem repeat DNA database for the human identity testing community. Nucleic Acids Res 29(1): 320-322.
- Tomas C, Sanchez JJ, Barbaro A, Brandt Casadevall C, Hernandez A, et al. (2008) X-chromosome SNP analyses in 11 human Mediterranean populations show a high overall genetic homogeneity except in North-west Africans (Moroccans). BMC Evolutionary Biol 8: 75.
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, et al. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29(1): 308-311.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12): 2725-2729.
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT, Nucleic Acids Symposium Series (Oxford) pp. 1979-2000.
- Huson DH (1998) SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14(1): 68-73.
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, et al. (2016) The Ensembl Variant Effect Predictor. Genome Biol 17: 122.
- Phillips C, Prieto L, Fondevila M, Salas A, Gómez Tato A, et al. (2009) Ancestry Analysis in the 11-M Madrid Bomb Attack Investigation. PLoS One 4: e6583.
- Hameed IH, Jebor MA, Ommer AJ, Abdulzahra AI, Yoke C (2016) Haplotype data of mitochondrial DNA coding region encompassing nucleotide positions 11,719-12,184 and evaluate the importance of these positions for forensic genetic purposes in Iraq. Mitochondrial DNA A DNA Mapp Seq Anal 27(2): 1324-1327.
- Santos C, Phillips C, Fondevila M, Daniel R, Van-Oorschot RAH, et al. (2015) Pacifiplex: an ancestry-informative SNP panel centred on Australia and the Pacific region. Forensic Sci Int Genet 20: 71-80.
- Wang Z, Moult J (2001) SNPs, Protein Structure, and Disease. Hum Mutat 17(3): 263-270.

 Research Article
Research Article