Abstract
Objective: Determining the frequency of MBLs classifying VIM, IMP and NDM in P. aeruginosa isolates.
Methods: The isolates of P. aeruginosa were isolated from sputum, blood, pus, fluid from patients (non-duplicate samples) in different clinical departments. They had been identified and performed antibiotic susceptibility testing by using Vitek 2 according to CLSI 2017 standards at Cho Ray Hospital from October 2017 to March 2018. The isolates were subcultured, picked colonies, extracted DNA and perform real-time PCR with MDR-MBL kit (VIM, IMP, NDM) of Sacace (Italy) to detect the MBL class VIM, IMP and NDM.
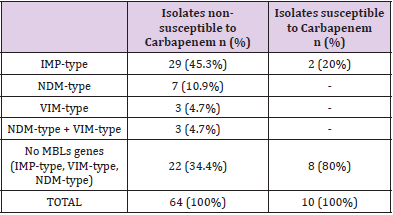
Results: There were 64 isolates of P. aeruginosa resistant to carbapenem (either or both Imipenem and Meropenem) were carried out real-time PCR to detect MBL (VIM, IMP, NDM). Among them, 29 (45.3%) isolates had IMP-type gene, 7 (10.9%) isolates had NDM-type gene, 3 (4.7%) isolates had VIM-type gene, 3 (4.7%) isolates had both IMPtype and NDM-type genes, 22 (34.4%) isolates were negative for the above 3 genes. 10 carbapenem-susceptible isolates were performed real-time PCR to detect MBL (IMPtype, NDM-type, VIM-type). Among them, 2 (20%) isolates had IMP-type gene, 8 (80%) isolates were negative for 3 genes.
Conclusion: The results of this study revealved a noticeably high prevalance of MBL- producing P. aeruginosa isolates at our hospital. MBL genes among isolates which reported as sensitive to carbapenem was acting as reservoir of such resistance genes with potential risk for the silent spread of these genes in hospitals and community. So, early recognition of MBL-producing isolates, establishing efficacy infection control measures and an appropriate protocol for antibicrobial therapy based on the laboratory data are necessary to prevent the dissemination of these resistance.
Keywords: Pseudomonas aeruginosa; MBL; IMP; NDM; VIM
Introduction
Background
Antimicrobial resistance is one of the biggest threat to global health especialy on developing country as Viet Nam. By 2050, if there is not effective interventions against antimicrobial resistance, 10 million deaths may occur annually [1]. P.aeruginosa is one of the most frequent and important pathogens in hospital-acquired infections, particularly in critically ill or immunocompromised patients, because they can be resistant to many common used antimicrobial agents. Carbapenems have been important antibiotics in MDR P. aeruginosa treatment. However, resistance to carbapenems has been reported, especially in developing countries [2]. The most common mechanism of resistance to carbapenems is the production of metallo-beta-lactamases (MBLs) [3]. MBLs are encoded either by genes on chromosome or on specific genetic elements so that they can resist to carbapenems and other antimicrobial agents, confer multidrug resistance (MDR) P. aeruginosa. In 1991, MBL producing P. aeruginosa were first reported in Japan, and since then there have been many reports of MBLs among carbapenem-resistant P. aeruginosa isolates in the world [4]. Mortality rates are signficantly higher in MBL producing P. aeruginosa than non–MBL P. aeruginosa [5]. Moreover, MBL can be spread from P. aeruginosa to members of Enterobacteriaceae, increasing the antimicrobial resistant rate and complication in the treatment of infected patients [6]. In addition, new antipseudomonal, such as ceftolozane-tazobactam, ceftazidime-avibactam, have better activity against MDR P. aeruginosa, however activity is just only in especially isolates with certain mechanism of resistance. Therefore, understanding the molecular characteristics of MBLs producing P. aeruginosa is very important to choose the appropriate treatment, control infection and prevent a possible global health crisis.
There are many different types of acquired MBL depending on epidemiological areas. The most important types that contribute to clinical studies are the IMP-type, VIM-type, NDM-type, SPM-type. However, at present, there is no standard phenotypic confirmatory tests for detection MBLs among P.aeruginosa isolates. [3] Several authors show that molecular methods is the proper test for screening the presence of MBLs genes Polymerase chain reaction (PCR) has high sensitivity and reliability for detection of MBL genes among MBL-producing P. aeruginosa. At present, there is no systematic evaluation of the molecular epidemiology of MBLproducing P. aeruginosa isolates in Viet Nam. Therefore, we carry on the study to detect the features of frequencies of IMP-type, VIMtype, NDM-type genes among P. aeruginosa isolates in our hospital.
Objectives
Due to the importance of detection MBLs, in this study we determined the frequency of MBLs classifying VIM, IMP and NDM in P. aeruginosa isolates in the patients by the PCR real-time method.
Materials and Methods
Bacterial isolates
This was a cross-sectional study, in which a total of 74 nonduplicated P. aeruginosa isolates were collected from various clinical samples received in the microbiology laboratory for bacterial culture and sensitivity from patients admitted in Cho Ray hospital at Ho Chi Minh city, located in the South of Viet Nam, during the period from October 2017 to March 2018. Bacterial species were identified using standard laboratory methods, including Gram staining, oxidase and on automated system such as Vitek 2-compact and MALDITOF-MS (Biomerieux, France). The isolates were stored at – 80oC in trypticase soy broth containing 20% glycerol and subculture twice prior testing.
Antimicrobial Susceptibility Testing
Antimicrobial Susceptibility Testing for all the isolates was done by automatic system Vitek2-compact to detect the minimum inhibitory concentrations (MICs). The following antibiotics were tested Piperacillin, Piperacillin/tazobactam, Ceftazidime, Cefepime, Gentamicin, Amikacin, Ciprofloxacin, Levofloxacin, Colistin, Meropenem, Imipenem. Susceptibility and MICs breakpoints were defined according to Clinical and Laboratory Standards Institute (CLSI; 2017). Carbapenem sensitivity was determined based on the MIC breakpoint. P. aeruginosa was defined non-susceptible to carbapenem when it’s inderteminate or resistant to imipenem and/or meropenem. P. aeruginosa ATCC 27853, E.coli ATCC 25922, E.coli ATCC 35218 were used as control isolates in all susceptibility assays.
Detection of MBLs by PCR
Carbapenem -non susceptible and carbapenem – susceptible isolates were tested by Sacace MDR MBL (VIM, IMP, NDM) Real-TM PCR kit. DNA was extracted by the SaMag™ Bacterial DNA Extraction Kit. Frozen bacteria were sub-cultured onto Mac-conkey agar (BD, American) before DNA extraction. International reference strain P. aeruginosa ATCC 27853 was used as the blaMBL gene’s negative control strain.
Results
74 isolates of P. aeruginosa from different clinical samples were collected. In total, 64 isolates were non-susceptible to these carbapenem, of which 42 (65.6%) isolates were found to be MBL producers. Twenty-nine (45.3%) isolates encoded IMP-type, which was the most commonly encoded gene among those investigated, followed by the NDM-type 7 (10.9%) and VIM-type 3 (4.7%), both IMP-type and NDM-type 3 (4.7%), and 22 (34.4%) isolates were negative for the above 3 genes. Ten carbapenem-susceptible isolates were performed real-time PCR to detect MBL, of which, 2 (20%) isolates had IMP-type gene, 8 (80%) isolates were negative for 3 genes IMP-type, VIM-type, NDM-type (Table 1).
Discussion
This discussion illustrated screening by molecular approach. In current study, 64 non-susceptible carbapenem isolates were collected. Among them, 42 (65.6%) isolates were MBL producers based on the PCR. This frequency was higher than in other studies at Asia or another region. In one report from Iran- 2017, prevalence of MBL-producing carbapenem-resistant P. aeruginosa was 43.7%. [7] In Thailand- 2019, of 153 carbapenem-resistant P. aeruginosa, this prevalence was 20.3% (31 isolates), divided into VIM-type (14 isolates) and IMP-type (17 isolates) [3]. This difference may be due to various research populations: Iran’s report was focus on the burn patients, and the Thailand’s report was based on the isolates from 5 regions of Thailand, our study was examined at our general hospital. This study found 29 (45.3%) isolates carried IMP-type, which was the most common MBL genes, followed by the genes NDM-type (10.9%) and VIM-type (4.7%). In 1988, transferable IMP-1 was isolated from P. aeruginosa in Japan and was found in a class 1 integrin located on a conjugational plasmid. Thereafter, it was identified in many other species suggesting horizontal gene transfer between unrelated Gram-negative species [4]. Therefore, in most studies, IMP type-producing isolates has a high prevalence. However, our result was different from a study in Iran and Thailand, that had equivalent frequency between VIM-type and IMP-type (17.5% and 15.6%) and (14 isolates and 17 isolates), respectively [8,9]. The difference may be due to the variation of geographical circulating strain.
In our present study, the NDM-type and VIM-type genes coexisted in 3 (4.7%) isolates. The genes encoding these enzymes are often located on transferable genetic platforms and can potentially spread to other species through horizontal gene transfer. Therefore, appropriate infection control guidelines and treatment protocols are needed to prevent the further dissemination of these resistant genes to other bacterial agents in hospital.
Unfortunately, we also detected 2 (20%) MBL-producing isolates (IMP-type) in carbapenem-susceptible P. aeruginosa. This result is similar to study in Sudan, from 2015 to 2016, prevalence of MBL genes by multiplex PCR among 100 carbapenems-susceptible gram-negative clinical isolates (including P. aeruginosa) was 27 (27%) [1]. It shows that, in some cases, MBL – producing isolates express low resistance, thus we cannot use the phenotypic test to detect them. This result has the important meaning, because MBL detection by PCR assays has not been routinely performed in most of clinical microbiology laboratories. Consequently, the occurrence of MBL production in P. aeruginosa that remains susceptible to carbapenem might be unrecognized by the clinical laboratory, they can transfer the genes to other gram negative because MBLencoding genes are often carried by mobile genetic elements, increasing the antibiotic resistance rate. In addition, failing in detection MBL production among carbapenem-susceptible isolates may lead to inadequate treatment, especially in serious ill patient.
Conclusion
The results of this study revealed a noticeably high prevalence of MBL- producing P. aeruginosa isolates at our hospital, especially in some carbapenem-susceptible isolates. MBL genes among isolates which reported as sensitive to carbapenem was acting as reservoir of such resistance genes with potential risk for the silent spread of these genes in hospitals and community. Thus, early recognition of MBL-producing isolates, establishing efficacy infection control measures and an appropriate protocol for antimicrobial therapy based on the laboratory data are necessary to prevent the dissemination of these resistance.
References
- Jim o Neil (2014) Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations.
- Cristina S, Andriana M, Pen C, Tubau F, Manzur A, et al. (2010) Influence of carbapenem resistance on mortality and the dynamics of mortality in Pseudomonas aeruginosa bloodstream infection. International Journal of Infectious Diseases 3: e73-e78.
- Franklin C, Liolios, Peleg AY (2006) Phenotypic Detection of Carbapenem-Susceptible Bacilli in the Clinical Laboratory 44(9): 3139-3144.
- Watanabe M, Iyobe S, Inoue M (1991) Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 35(1): 147-151.
- Lu A, Zavascki AP, Luı A (2006) The influence of metallo- b -lactamase production on mortality in nosocomial Pseudomonas aeruginosa infections. (June) 387-392.
- Peleg AY, Franklin C, Bell JM (2005) Dissemination of the Metallo- b -Lactamase Gene bla IMP-4 among Gram-Negative Pathogens in a Clinical Setting in Australia (October): 1549-1556.
- Azimi A, Peymani A, Kianoush P (2018) Major Article Phenotypic and molecular detection of isolates from patients with burns in Tehran , Iran 51(5): 610-615.
- Adam MA, Elhag WI (2018) Prevalence of metallo-β-lactamase acquired genes among carbapenems susceptible and resistant Gram-negative clinical isolates using multiplex PCR, Khartoum hospitals, Khartoum Sudan. BMC Infect Dis 18(1): 4-9.
- Khuntayaporn P, Yamprayoonswat W (2019) Dissemination of Carbapenem- Resistance among Multidrug Resistant Pseudomonas aeruginosa carrying Metallo-Beta-Lactamase Genes , including the Novel bla IMP-65 Gene in 51(2): 107-118.

 Research Article
Research Article