Abstract
Smart FlareTM RNA Detection Probes from Millipore is a novel technology to detect RNA in live cells based on the use of 12 nm gold nanoparticles coated with nucleotides. We proved that SmartFlaresTM are internalized by human primary lymphocytes. However, fluorescence signals from target RNA detection can only be observed in the presence of Fetal Bovine Serum (FBS) in the medium, whereas it is not detectable without FBS or when medium is supplemented with human albumin. Image analysis of fluorescence generated from SmartFlare™ Uptake Control (gives constant signal regardless of contact with RNA) and RNA Specific Probes revealed further differences. In the presence of FBS, the fluorescence signal for both reagents was diffused within the cells, whereas in the absence of FBS, it was detected as single spots within the cells only when the Uptake Control was used. It is possible that FBS components are necessary for SmartFlare™ Probes to be released from cellular compartments into the cytoplasm where they can get into contact with target RNA. The exact mechanism of this phenomena should be further determined. However, for the first time, we present here that FBS in the cell culture medium is essential for RNA detection by SmartFlare™ technology in human lymphocytes.
Keywords: Smart Flares™; Gold Nanoparticles; RNA Detection in Live Cells; Cellular Uptake
Introduction
Smart FlareTM RNA Detection Probes is a recently introduced platform described as the first known technology allowing for gene expression detection and quantification in live cells. The platform was developed by Mirkin’s group and commercialized by EMD Millipore [1]. The SmartFlareTM technology uses spherical gold nanoparticles covered with oligonucleotides, which are coupled single-stranded DNA. One of the strand is designed to be complementary to target RNA [2] and the shorter one has a fluorophore attached. The fluorescence signal from the fluorophore is efficiently quenched until it is in the proximity of the gold nanoparticle [3]. The SmartFlareTM particles are naturally internalized by live cells. Inside the cells, oligonucleotides on the nanoparticles bind to complementary target RNA, causing the release of the shorter DNA strands with fluorophore [1,2,4]. SmartFlareTM is an attractive tool for gene expression analysis in living cells due to its unique ability to enter live cells, lack of cytotoxicity and simplicity of application. Furthermore, the possibility to detect RNA in live cells makes it feasible to separate one cell type from another based on RNA expression via Fluorescence Activated Cell Sorting (FACS) and to use live cells in downstream applications. Several scientific studies have reported successful use of SmartFlaresTM for detection of specific RNA in multiple cell types including stem cells [5], myocytes [6], various cancer cell types [7,8] and monocytes [9]. However, there are still some controversies regarding SmartFlareTM technology.
For successful detection of target RNA and release of fluorophore, the nanoparticles must enter the cell and get into contact with the cytosol, where target RNA is localized. The mechanism, by which SmartFlaresTM enter cells is not entirely known, but most likely, they are naturally engulfed by cells via endocytosis [10]. In order for SmartFlareTM Probes to serve as a detection tool upon cell entry, the nanoparticles should be localized in cytoplasm to be able to get into contact with the target RNA. This is somewhat controversial, because other studies have suggested that gold nanoparticles remain entrapped in endo-lysosomal vesicles and never reach the cytoplasm within the cell [11,12]. Therefore, it is important to test if SmartFlareTM technology can be used for RNA detection and to define the factors that can influence successful SmartFlares™ application. Here, we present our results after testing SmartFlaresTM on human primary lymphocytes. During the experiments, we found that both the presence and type of serum in the cell culture medium play a crucial role in the SmartFlareTM uptake and RNA detection processes.
Materials and Methods
Isolation and Preparation of Cells
Primary human T cells were isolated from the fresh whole blood obtained from the healthy donors using RosetteSep™ Human T Cell Enrichment Cocktail (Stem Cell Technologies, Vancouver, BC, Canada) according to the instructions provided by the manufacturer. They were washed twice with Phosphate Buffer Saline (PBS; HyClone, Logan, UT, USA) and then counted after staining with 0.4% Trypan Blue (Amresco, Solon, OH, USA) for exclusion of non-viable cells. Isolated T cells were re-suspended in CTS™OpTmizer™ T Cell Expansion Medium supplemented with 26 mL/L of CTS™OpTmizer™ T Cell Expansion Supplement (Life Technologies, Grand Island, NY, USA) as recommended by the manufacturer, containing different concentrations (0.5, 1, 2%) of Fetal Bovine Serum (FBS; HyClone, Logan, UT, USA) or 2% of human serum albumin (HA; Flexbumin 25%, Baxter, Deerfield, IL, USA). T cell suspensions in each type of medium were prepared at concentration 106 cells per 1 mL of medium. After suspension preparation, 100 μL was added per one well of 96-well flat bottom plate. (Cells treat, Pepperell, MA, USA). Cells were cultured for up to 6 hours at 37oC, 95% humidity, 5% CO2 incubator until the preparation of SmartFlares™.
Preparation of Smartflares™ and Staining of Cells
The following controls and probe were used: SmartFlare™ Scramble Control for specificity (measures level of the background fluorescence), SmartFlare™ Uptake Control (checks, if the nanoparticles can enter the cells) and SmartFlare™ 18S RNA Probe (detects specific target RNA of housekeeping gene - 18S). All Controls and the Probe were conjugated with Cyanine-3 (Cy3) fluorophore. Stock solutions of Controls and the Probe were prepared as per manufacturer instructions by dissolving lyophilized powder in 50 μL of nuclease-free water (Life Technologies, Carlsbad, CA, USA). Dissolved stock solutions of controls and the probe were kept in the dark at room temperature. Working solutions were prepared by diluting the stock solutions 20x in PBS. The reagents were added drop wise into the designated wells with cells for each of the Controls or the Probe at final concentration of 666 pM. Then, medium with different concentrations of FBS/HA were added to the wells, up to the final volume of 200 μL per well. Cells with SmartFlare™ Controls or the Probe were incubated at 37oC, 95% humidity, 5% CO2 incubator for 16h.
Cell Viability after Incubation with SmartFlares™
To perform cell viability analysis of cells incubated with SmartFlares™, we used LIVE/DEAD® Fixable Green Dead Cell Stain Kit (Molecular Probes, Inc., Eugene, OR, USA). T cells and SmartFlare™ RNA Uptake Control conjugated with Cyanine-5 (Cy5) were prepared in the same fashion as described above for the rest of experiments. After a 16h incubation, T cells were collected, washed with PBS, and stained with LIVE/DEAD® Fixable Green Dead Cell Stain reagent for 15 minutes as indicated in the instruction provided by the manufacturer. Cells were analyzed by flow cytometry (LSR II Cell Analyzer, BD Biosciences, San Jose, CA, USA). The non-viable cells were identified, gating the cell population with the highest fluorescence emission at 530 nm.
Detection of Signal from Smartflares™ using Flow Cytometer
After incubation time, cell suspensions from each condition were collected separately and washed once with PBS. Pellets were re-suspended in 400 μL of PBS and transferred immediately for flow cytometric analysis. A minimum of 5,000 events were acquired per analyzed sample on LSR II Cell Analyzer (BD Biosciences, San Jose, CA, USA).
Visualization of Smartflares™ RNA Probes in Cells using Amnis Imagestream-X
Cells incubated with Cy3 labeled SmartFlare™ Uptake Control in the absence and presence of FBS or with Cy3 labeled SmartFlare™18S Probe in the presence of FBS were collected and washed once with PBS. Pellets were re-suspended in 60 μL of PBS and transferred immediately for analysis on Amnis ImageStream® X (EMD Millipore Corp. Darmstadt, Germany). A minimum of 1,000 events were acquired per analyzed sample.
Data Analysis and Statistics
Flow cytometric data were analyzed on FlowJo® software (FlowJo, LLC, Ashland, OR, USA). Fluorescence of the Negative Control (cells without addition of SmartFlares™) was used for gate settings. Statistical analysis was performed with the use of GraphPad Prism v5.0 software (GraphPad Software, Inc., San Diego, CA). Data are expressed as mean +/- SEM.
Results
Serum in the Medium does not Affect Smartflare™ Nanoparticles Uptake but is Critical for RNA Detection
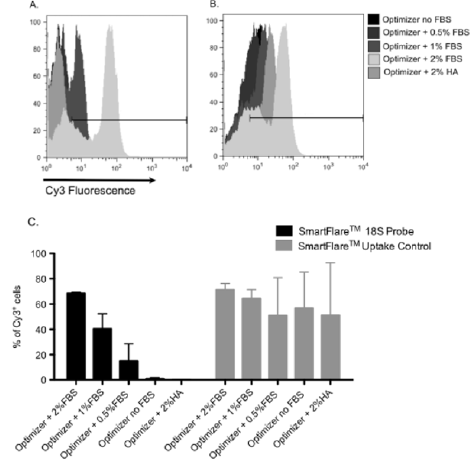
In order to optimize protocol for RNA detection in human primary lymphocytes using SmartFlare™ Probes, we initially used SmartFlare™ Uptake Control. When using this control, a fluorescence signal could be observed, regardless of contact with RNA. Therefore, it can be used for measurement of the rate of SmartFlares™ uptake by cells. After the 16-hour incubation of isolated human lymphocytes with SmartFlare™ Uptake Control, an average of 60% of all cells were Cy3-positive, regardless of the FBS concentration used in the medium for cell incubation (Figure 1). This indicates that SmartFlare™ nanoparticles were able to enter the cells. We also checked, if SmartFlare™ nanoparticles affected cell viability. After the T cells were incubated for 16-hours with SmartFlare™ Uptake Control Probe, the average percentage of viable cells was 94.4%, indicating that cell viability was well preserved. In the next step, we used the SmartFlare™ 18S rRNA Probe to confirm that gene expression detection is possible by SmartFlares™. The Cy3 fluorescence from 18S SmartFlare™ Probe was detected in an average of 68% of all T cells, when medium with 2% FBS was used for incubation of cells with the probe. The frequency of Cy3+ cells decreased to 40% and 14%, when a medium with 1% and 0.5% FBS was used, respectively (Figure 1). In the absence of FBS in the medium, no cells indicated fluorescence signal upon incubation with SmartFlares™ 18S rRNA Probe (Figure 1). No labeled cells were also detected, when medium was supplemented with 2% Human Serum Albumin instead of complete FBS (Figure 1). These findings suggest that the detection of the signal from SmartFlare™ Probes is associated with specific factors present in FBS, which are different than albumin.
Figure 1: Impact of medium supplementation on cellular uptake of SmartFlare™ nanoparticles and 18S RNA detection by SmartFlares™ in human T cells. Isolated human T cells were incubated with Cy3 labeled SmartFlare™ 18S Detection Probe or SmartFlare™ Uptake Control in the presence of FBS at different concentrations (2%, 1%, 0.5%), absence of FBS and 2% HSA. Sets of histograms from representative flow cytometric analysis are showing Cy3 fluorescence of the cells after incubation with SmartFlare™ 18S Probe (A) and SmartFlare™ uptake control (B). The gate was set on cells incubated without SmartFlares™. Percentage of Cy3 positive cells is shown in chart (C). The results shown are mean ± SEM, n=3.
In the Absence of Serum Signal from the Smartflares™ Localizes in Vesicles
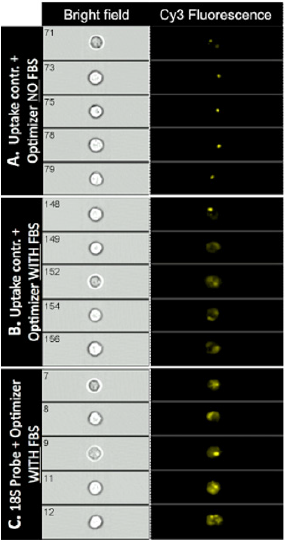
In order to further investigate cellular uptake of SmartFlare™ nanoparticles and the effect of FBS on RNA detection by SmartFlares™, we performed analysis on ImageStream® Imaging Flow Cytometer. This instrument captures high-resolution bright field and fluorescence microscopic images of every cell analyzed in the stream. When the cells were incubated with SmartFlares™ without FBS, the signal from the Uptake Control was localized in the spherical areas within the single cells, whereas in the presence of FBS in the medium, the signal was dispersed and not limited to a spherical-shape (Figure 2). Similarly, when detecting 18S RNA with SmartFlares™ in the presence of 2% FBS in the cell culture medium, a fluorescence signal was dispersed in the cells (Figure 2). After incubating cells with SmartFlare™ 18S Probes in the medium without serum, we did not observe spread or spherical fluorescence (data not shown).
Figure 2: Visualization of SmartFlare™ nanoparticles uptake and 18S RNA detection by SmartFlares™ on single cell level after cell incubation in presence or absence of FBS. Human T cells after isolation were incubated with Cy3 labeled SmartFlare™ Uptake (A,B) or with Cy3 labeled SmartFlare™ 18S Detection Probe (C). Cells with SmartFlares were incubated in medium without FBS (A) or supplemented with 2% FBS (B, C). After incubation, cells were collected, washed and analyzed on the ImageStream® X. In the first column on the left (Bright field), single cells were visualized under the bright field. The second column (Cy3 Fluorescence) shows detection of Cy3 Fluorescence within the single cells. Cy3 fluorescence signal from the SmartFlares™ Uptake Control was localized in the cell’s spherical areas when no FBS was added in the medium during T cells incubation with nanoparticles (A). In the presence of FBS in medium, the signal was dispersed for SmartFlare™ Uptake Control (B) and for the SmartFlares™ 18S RNA Detection Probe (C). Data are shown from the one representative experiment.
Discussion
Here, we are showing for the first time that human lymphocytes are capable of internalizing oligonucleotide-covered nanoparticles (SmartFlares™). After optimization of experimental conditions like concentration of the SmartFlare™ nanoparticles, density of the cells and incubation time (data not shown), we were able to detect a fluorescence signal using the SmartFlare™ Uptake Control within isolated T cells. As part of the optimization experiments, we also tested whether a FBS supplement added to the culture medium can affect or contribute to the cellular uptake rate of SmartFlare™ nanoparticles. We found that T cells are able to uptake SmartFlares™ as long as the medium was supplemented with FBS. There was no flouorescence signal detected by SmartFlare™ Probes when checking 18S housekeeping gene expression with this technology without presence of FBS in medium. Additional testing revealed that with the higher concentration of FBS in the medium, there is an increase in the percentage of T cells presenting fluorescence signals from Cy3 dye. This observation indicates that detection of 18S ribosomal RNA by SmartFlare™ Probes may be efficient, but only when FBS is present in the medium during incubation of the cells with the probes. Furthermore, ImageStream® analysis revealed that the fluorescence signal from the Uptake Control and 18S Detection Probe in the presence of FBS was dispersed within the cell. However, when the cell culture medium was not supplemented with FBS, the fluorescence signal was presented in spherical spots for the SmartFlare™ Uptake Control and undetected for the SmartFlare™ 18S Probe.
These observations are indicating that some components of FBS are critical for RNA detection by SmartFlare™ Probes but are not essential for the nanoparticles to enter the cells. It is possible that SmartFlare™ nanoparticles can be successfully internalized by cells in the absence of FBS, but they might become trapped in intracellular compartments and are not able to get into the contact with target RNA. Such types of observations have already been described. In 2014 Wu et al. reported that only a small percentage of gold nanoparticles coated with fluorescently labeled nucleic acids reach the cytoplasm and most of them remained trapped in the endosomes. Moreover, the results of that work indicate that oligonucleotide-covered nanoparticles in the late endosomes, are subjected to DNase activity, which cause cleavage of the nucleotides from their surface [11]. In this scenario, detection of target RNA by oligonucleotide- covered gold nanoparticles also failed. The results of another group were similar. Oligonucleotide-covered gold nanoparticles were able to enter cells, but they remained trapped in the endocytic compartments and were not able to serve as probes to detect cytoplasmic RNA [12].
In a similar technology, called Sticky-flares, designed to track RNA particle movement in the live cells, a similar observation was reported [13]. Sticky-flare technology is based on the same concept as the SmartFlares™ with nucleotide-covered nanoparticles that can enter the cell, bind to target RNA, and release fluorophores [13]. The only difference with Sticky-flare is that the oligonucleotide strand complementary to the target mRNA is detached from the nanoparticle, and after hybridization, can be used for RNA tracking within the cell [13]. In one of the comments in the Sticky-Flare article discussion, there is speculation about the fluorescence signal localization in the cells observed in the article’s figures [14]. In their opinion, the signal comes from endosomes rather than other cell compartments where RNA could be present [12,14].
In summary, based on our experiments optimizing the conditions for SmartFlares™ to detect RNA in human lymphocytes, we concluded that:
a) Human primary lymphocytes are able to uptake SmartFlare™ nanoparticles.
b) FBS used as a cell culture medium supplement for suspending and incubating cells with SmartFlare™ Probes has a beneficial effect on RNA detection.
c) FBS has no effect on SmartFlare™ nanoparticle cellular uptake, but it is crucial for RNA detection.
d) Supplementation of the cell culture medium with human albumin does not allow for RNA detection via SmartFlare™ nanoparticles, thus the effect of serum is component-specific.
We are aware that further experiments are needed to better understand the exact mechanism of the FBS effect on RNA detection by SmartFlares™. Nevertheless, our observation that the presence of FBS in the cell culture medium is critical for the successful application of SmartFlares™ has a practical value for researchers from many different fields. Utilizing SmartFlare™ technology more efficiently will save time and resources, progressing their projects faster. Therefore, we decided to share our results in the preliminary phase without defining the exact mechanism and components responsible for the effect.
Acknowledgement
We would like to also thank Dean Kenneth Polonsky and Dr. Jeffrey Matthews, Chair of the Department of Surgery at The University of Chicago for their generous support of the project.
Compliance with Ethical Standards
Funding: This work was funded by
a) US Public Health Service Grant DK-020595 to the University of Chicago Diabetes Research and Training Center,
b) CRC- National Center for Advancing Transitional Sciences of the NIH Grant # UL1TR000430,
c) National Centre for Research and Development, Poland (grant no STRATEGMED1/ 233368/1/NCBR/2014,
d) National Science Centre, Poland (decision no Dec2011/03/D/ N25/00555)
e) The Foundation for Polish Science (grant nr 43/UD/ Skills/20150) for Adam Krzystyniak.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Prigodich AE, Randeria PS, Briley WE, Kim NJ, Daniel WL (2012) Multiplexed nanoflares: MRNA detection in live cells. Anal Chem 84(4): 2062-2066.
- Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA (2007) Nano-Flares: Probes for Transfection and mRNA Detection in Living Cells. J Am Chem Soc 129(50): 15477-1579.
- Dubertret B, Calame M, Libchaber AJ (2001) Single-mismatch detection using goldquenched fluorescent oligonucleotides. Nat Biotechnol 19(4): 365-370.
- Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC (2009) Nano-flares for mRNA Regulation and Detection. ACS Nano 3(8): 2147-2152.
- Lahm H, Doppler S, Dreßen M, Werner A, Adamczyk K, et al. (2015) Live fluorescent RNA-based detection of pluripotency gene expression in embryonic and induced pluripotent stem cells of different species. Stem Cells Dayt Ohio 33(2): 392-402.
- Mehta A, Sequiera GL, Ramachandra CJA, Sudibyo Y, Chung Y, et al. (2014) Re-trafficking of hERG reverses long QT syndrome 2 phenotype in human iPS-derived cardiomyocytes. Cardiovasc Res 102(3): 497-506.
- Seftor EA, Seftor REB, Weldon DS, Kirsammer GT, Margaryan NV (2014) Melanoma tumor cell heterogeneity: a molecular approach to study subpopulations expressing the embryonic morphogen nodal. Semin Oncol 41(2): 259-266.
- McClellan S, Slamecka J, Howze P, Thompson L, Finan M (2015) mRNA detection in living cells: A next generation cancer stem cell identification technique. Methods 82: 47-54.
- Khare S, Ratsimandresy RA, de Almeida L, Cuda CM, Rellick SL, et al. (2014) Nat Immunol 15(4): 343-353.
- Ma D (2014) Enhancing endosomal escape for nanoparticle mediated siRNA delivery. Nanoscale 6(12): 6415-6125.
- Wu XA, Choi CHJ, Zhang C, Hao L, Mirkin CA (2014) Intracellular fate of spherical nucleic acid nanoparticle conjugates. J Am Chem Soc 136(21): 7726-7733.
- Comenge J, Held M, Levy R, Carolan G, Mason D (2015) The Spherical Nucleic Acids mRNA Detection Paradox.
- Briley WE, Bondy MH, Randeria PS, Dupper TJ, Mirkin CA (2015) Quantification and real-time tracking of RNA in live cells using Sticky-flares. Proc Natl Acad Sci 112(31): 9591-9595.
- Mason D, Levy R (2015) Sticky-flares: real-time tracking of mRNAs... or of endosomes? bioRxiv. 19: 029447.

 Research Article
Research Article