Abstract
During fetal growth and development, placental integrity is of the utmost significance, which determines the success of pregnancy. The placenta is an exceedingly specialized transitory organ that is precisely regulated by endocrine, metabolic and immunological processes. It develops from the blastocyst in a manner that resembles a growing tumor through cellular invasion and angiogenesis but unlike tumor, these processes are strictly controlled. The rapid and undifferentiated growth of the trophoblast gives rise to the villous and the extravillous phenotypes, which differentiates together in order to establish fetal-maternal circulation. While the villous lineage enters the syncytial pathway, the extravillous trophoblast invades the maternal decidua. The aim of this review is to unravel imprinted gene regulation in the placental and fetal development, and their apparent effects on the future health of the fetus using transcriptomic and proteomic approaches. Understanding the genetic influence of placental development and function is of great clinical interest since any variation may lead to pregnancy related complications such as preeclampsia.
Keywords: Gene Regulation; Placenta; Pregnancy; Fetal Development; Preeclampsia
Introduction
The development of placenta into the various trophoblast subtypes helps to determine the success of a pregnancy outcome. The human placenta fulfills different functions such as enabling the blastocyst adaptation in utero and nourishment of the developing fetus. Impairment in placentation in early gestational stages is implicated with a number of conditions such as preeclampsia, miscarriages and restriction of intrauterine growth, which may negatively affect the health of the developing fetus or even the future of the child through fetal programming. Previous studies indicate that during the first semester, system genes that are associated with biological mechanism including nucleic acids and protein metabolism, mRNA transcription and other cellular processes are highly expressed and regulated. This indicates the intensity of cellular proliferation and differentiation during this gestational stage. These findings have been reported by Mikheev, et al. [1] as well as other genomic studies on placental development and function [2]. Synthesis in the placenta secretes large amount of molecules that are essential for its development, metabolism and fetus growth. The factors involved include hormones of the placenta and growth factors which lead to the regulation of gene expression critical for plasticity of the placenta plasticity and functions. Placental investigation requires an approach which is high throughput and thorough methods for example microarray, ribonucleic acid sequencing (RNA-Seq) and/or quantitative reverse transcription-polymerase chain reaction (qRT-PCR) technologies, in tissue analysis for physiology and pathophysiology.
Expression of this transcript correlates with the maternal blood pressure as well as neonatal birth weight, which suggest the role it plays in pre-eclampsia. These collective reviews for imprinted gene regulation signatures might be used in future as new biomarkers for maternal and fetal complications of pregnancy. In this review, literature search was performed in the last fifteen years in PubMed and Google scholar, using the key terms: placental gene expression / regulation and fetal development. Only published articles with a set gene involved in normal placental function and pathogenesis were considered. We chose recently published to get the true picture of Imprinted Gene Regulation on Placental and Fetal Development, but also cited relevant and important older publications where necessary. Currently there is inadequate information on the effect of gene expression and regulatory mechanisms on placental and fetal development, this review, therefore, will shed more light on gene expression involved in placental and fetal development as well as identifying the areas that needs further studies.
Unique Patterns of Gene Expression in the Placenta
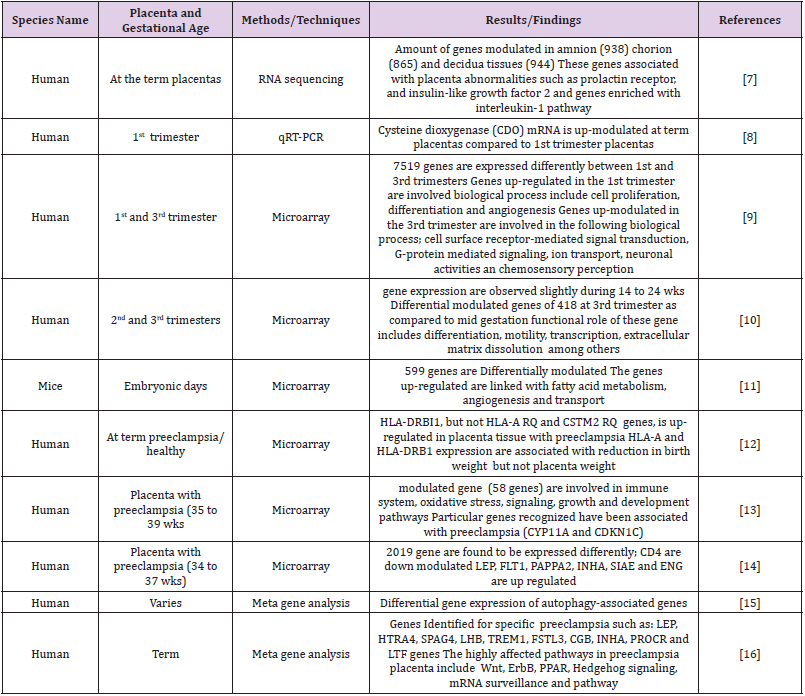
Various mRNA transcripts such as human chorionic gonadotropin β subunit (hCGB), corticotropin-releasing hormone (CRH) and human placental lactogen (PL) have been characterized in maternal plasma [3]. The expression pattern of each gene is dependent on the gestational age. Transcripts such as fetal-derived γ-globin increase in maternal circulation after elective pregnancy termination [4]. Placental transcripts of clinical significance have been characterized through microarray analysis to generate noninvasive fetal gene profiles [5] including the increased CRH levels in preeclampsia [6] and the decreased HS3ST3A1 mRNA expression in pre-eclamptic placental tissue. The imprinted gene regulation of placenta and fetal development during different gestational stages is summarized in Table 1. Additionally, identified common gene pathways involved in imprinted gene regulation of placental and fetal development are also shown. In mammals, genetic imprinting is a significant process where many genes undergoes epigenetic modification and imprinted specifically in the placenta [7-18]. Placental imprinting appears to be a continuous process that occurs throughout the pregnancy [19]. Data from the entire genome methylation from placental tissue during the first and third trimester indicates a methylation-induced down regulation for a number of tumor related genes as a normal placentation process [20]. In this review, it is reported imprinted genes are expressed in a temporal process in the course of normal human placenta development. The study has also demonstrated differential gene expression of PHLDLA2 and IGF2 between first and third trimester of placental tissue. Additionally, gene expression in the placenta is also affected by metabolic conditions such as maternal obesity and gestational diabetes mellitus (GDM) by affecting the energy sensing, which modulates the maternal body mass index (BMI) and GDM on birth weight.
Table 1: Gene expression and regulation during placental and fetal development.
Note: CDO, Cysteine dioxygenase; HLA-DRBI1, Human Leucocytes Antigen, DRB1 beta chain; CYP11A, Cytochrome P11A; CDKN1C, Cyclin Dependent Kinase Inhibitor 1C; LEP, Leptin; FLT1, Fms Related Receptor Tyrosine Kinase 1; PAPPA2, Pappalysin 2; INHA, Inhibin, alpha; SIAE, Sialic Acid Acetylesterase; ENG, Endoglin; HTRA4, high‑temperature requirement A 4; SPAG4, Sperm Associated Antigen 4; LBH, Luteinizing hormones beta-subunit; TRM1, Triggering Receptor Expressed On Myeloid Cells 1; FSTL3, Follistatin Like 3; CGB, Chorionic gonadotropin beta; PROCR, Protein C Receptor; LTF, Lactotransferrin.
During early human gestation, there is a high expression of ATP Binding Cassette Subfamily B Member 1 (ABCB1) mRNA and P-glycoprotein (P-gp protein), which could remarkably reduce near term [21,22]. However, there are inconsistent findings on the gestational expression of breast cancer resistance protein (ABCG2)/ATP-binding cassette subfamily G member 2 (BCRP) in human placenta [23,24]. While some studies have findings showing no change in expression, others have reported a significant increase or decrease in expression [25-27]. The inconsistent findings are attributed to the limited number of placental tissues used in the studies or even the sample quality. Only a few studies have explored the relative expression of drug transporters in human placental tissue. Some of the studies report a high expression of BCRP compared to that of P-gp in primary term trophoblast cells [28]. There is a remarkable reduction in P-gp levels at term labor but the relative expression of P-gp and BCRP during early stages of pregnancy is yet to be understood. There is also limited understanding on the regulation of placental ABCB1/Pgp and ABCG2/BCRP but a number of regulatory elements and transcription factors of ABCB1 and ABCG2 gene promoters have been reported to be significance in gene activation.
Placental RNA Quality and Quantity
Evaluation of the quality and quantity RNA is essential for analysis of gene expression; this is reflected in degraded samples which might influence the interpretation of RNA expression levels. Study by Monk et al., [19] on imprinted genes specific to human placenta taken from first and third trimester placental tissue along with maternal and third trimester paternal blood samples was largely by biallelic gene expression (cyclooxygenase 2 and 5B (COX2, COX5B) and cytochrome P450 (CYP) 2D1 and −2D7 isoforms (CYP2D1, CYP2D7)) throughout the gestational period. Validation of these genes by PCR reported a specific regulation of COX2 and COX5B in cases with smoking history regardless of gestational age. Studies report that there are epigenetic modifications that may directly influence the size; morphology as well as transport capacity of the placenta [20]. Samples of Placental tissue from preterm and term deliveries alleged to be from preterm labor presented a significant increase in expression of TNFα and IL6, with decreased expression of IFNγ [29]. The placental epigenetic status can be influenced by environmental factors that may in turn affect the fetal growth and development [21].
Gene Annotations
For gene annotations, Protein Analysis Through Evolutionary Relationships (PANTHER) [30] software system has been used to analyze gene sequence and relate them to their particular biological processes and molecular roles (http://www.pantherdb.org). The signaling pathways involved during normal placental development have been identified using Bonferroni correction for multiple testing and expression data analysis tool [9]. Analysis of Pathways of differential gene expression between first and third trimester of the placentas reported angiogenesis to be extremely active in first trimester.
Placental Gene Imprinting
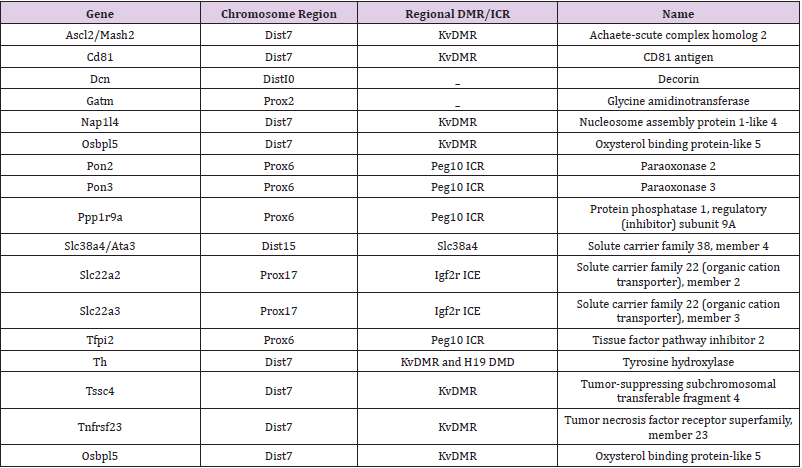
Fetal growth and development of the placenta are regulated by imprinted genes in mammalian species and are thought to have co evolved with placentation (Table 2). In majority of mammals, autosomal genes are expressed co-dominantly from the two parental chromosomes. The process of monoallelic expression is attained through epigenetic asymmetry between parental alleles such as maternal gene expression specific to the placenta and are imprinted regardless of their elevated level of expression in decidua as in the case of Tissue Factor Pathway Inhibitor 2 (Tfpi2) [31]. As a result, false-positive maternal expression due to decidua contamination can be distinguished from actual imprinting by making use of in situ staining or from backcrosses whereby homozygous embryos are formed in a heterozygous mother. In such cases, the allele detected can be absent in the embryo’s genome which signify maternal contamination [32]. Placenta contains several specific imprinted transcripts, which are mainly found in the large imprinted domains as shown in Table 2. For instance, two clusters of maternally expressed genes specific to the placenta are located on the distal chromosome seven and proximal chromosome seventeen, both of which are regulated by maternally methylated regions, the KvDMR1 and Airn DMRs, respectively [33,34]. In these DMRs, there are promoters for long ncRNAs. The Airn non coding RNA (ncRNA) transcript silences two adjacent genes, solute carrier family 22 member 2 (SLC22A2) and SLC22A3 in the Igf2r domain [35] through recruitment of histone methyltransferase enzymes such as G9a to the paternal allele [36], and deposit of repressive histone mark on lysine 9 of histone H3 (H3K9me2) [37]. Additionally, Airn cause direct imprinting of Igf2r gene since transcription via the Igf2r promoter on the paternal allele is adequate for silencing, probably by dislocating transcription factors important for Igf2r expression [9]. The long paternal expression of ncRNA KCNQ1OT1 on chromosome seven recruit G9a and together with H3K27me3 histone methyltransferase enhancer of zeste homolog 2 (Ezh2), results to the paternal silencing of eight flanking genes within the placenta [38]. Interestingly, within the KCNQ1OT1 loci, imprinting is not conserved in human placenta due to lack of allelic repressive histone modifications. The imprinting of SLC22A2 and Insulin-like growth factor 2 receptor (IGF2R) is polymorphic in humans [39]. The role of these genes in placenta-related complications and intrauterine growth restriction should further be investigated along with aberrant genes as biomarkers in complicated pregnancies, in order to help in the diagnosis of at-risk pregnancies during early gestation [40].
Placental Gene Regulation
Certain genes in the placenta are under strict epigenetic regulation and therefore prone to genomic imprinting. The variation in epigenetic profiles and allelic expression of imprinted genes in different species reflects the evolutionary dynamic and adaptive phenomenon under epigenetic control [41]. In fetal growth restriction (FGR) the complications with absent end-diastolic blood flow (AEDF) in umbilical artery, the expression of neuropilin 1 (NRP-1) placental is down regulated. NRP-1 plays an important role in angiogenesis and its down regulation may result into lack of sufficient vascular branching as in the case of FGR placental complications that portray an antiangiogenic state [42]. There is a need for further research in order to elucidate the mechanisms involved or to develop new diagnostic and therapeutic tools. Based on various studies, the computation of standardized mean difference (mu) of the six genes exhibit a differential expression in the initial meta-signature two of the twelve genes (VIM and HSD17B1) were amongst the 688 genes that were differentially expressed, but they were left out during the leave-one-out-analysis [43]. Four genes (Insulin like growth factor Binding protein 1 [IGFBP1], chromogranin A [CGA], TNFSF10/TRAIL and PGF) had no significant differential expression based on the meta-analysis of the original microarray data [44]. The differential expression of genes included in gene list-driven signature in the current analysis, their profiles were similar except for subtilis phage pavia 1 (SPP1), superoxide dismutase (SOD1), and Verona integron-encoded metallo-lactamase (VIM). Studies indicate up regulation and down regulation of SOD1 and VIM in the pre-eclamptic placenta and in the current analysis, they are down regulated. In the pre-eclamptic placenta, SOD1 is one of significantly down regulated genes [45].
Hypoxia/ischemia compromise pre-eclamptic placentas and hypoxia-inducible factor-1 alpha (HIF-1α) mainly mediate gene expression. Over 10% of the meta-signature genes that are up regulated are directly targeted by HIF-1α CREB binding protein (CREBBP)/EP300, which is a key transcriptional co activator of HIF-1α, therefore, low CREBBP/EP300 level may reduce the placenta’s ability to respond to shortage of oxygen, which worsen the pre-eclamptic state [43]. A study from previous work indicated the concentration of tumor necrosis factor (TNF-α), interleukin 1 alpha (IL-1β) and IL-6 in serum was higher in GDM group compared to the control group. Forkhead box O1 (FoxO1) expression was detected in adipose tissue of both the placenta and fetus. When compared to the control group, the gene and protein expression of FoxO1 & TNF-α was higher in the GDM group in both tissues [46]. Another study also reported a positive co-relation between FoxO1 expression in the placenta with homeostatic model assessment of Insulin Resistance (HOMA-IR) and TNF-α. TNF-α gene stimulation increases FoxO1 expression in trophoblast cell cultures. Deletion of FoxO1 in the cells reduces TNF-α-induced expression of IL-6 and IL- 1β pro-inflammatory cytokines [47]. These findings indicate that FoxO1 plays as a pro-inflammatory factor in GDM as well as in IR by interacting with TNF-α, a pro-inflammatory cytokine that helps to modulate the acute phase reaction and first discovered in placenta and amnion [37]. The insulin-like growth factor-binding protein (IGFBP) also known as pregnancy-associated plasma protein A2 (PAPPA2), protease that is highly expressed in the placenta is up regulated in pre-eclampsia; HELLP (Haemolytic anaemia, Elevated Liver enzymes, and Low Platelet count) syndrome [48].
High expression of PAPPA2 results to abnormal placental development and its up regulation may be meant to compensate for placental pathology. Oxidative stress and hypoxia are among other conditions that affect the expression of PAPPA2 in preeclamptic placenta; TNF-α and prostaglandin E2 (PGE2) results to the up regulation of Pappalysin 2 (PAPPA2) [49]. Hypoxia, which is common in pre-eclamptic placenta results to PAPPA2 expression. These findings confirm the hypothesis of up regulation of PAPPA2 as a result of placental pathology as opposed to the assumption that high PAPPA2 levels cause preeclampsia [50]. The aryl hydrocarbon receptor (AhR) and peroxisome proliferators-activated receptor gamma (PPARg) are also reported to regulate ABCG2/BCRP and ABCB1/P-gp [51]. However, the role of transcription factors such as p53 and AP-1 on the expression of placental drug transporter genes requires further elucidation.
Placental DNA Methylation
Extensive research has been done on the effect of imprinted methylation on a number of placental abnormalities stated in the DNA methyltransferase 1o (DNMT1o) model, initially, DNA methylation determined on fifteen imprinted gametic differentially methylated domains (gDMDs) is thrice during the last half of gestation period [52]. The mean methylation fraction across twelve non-redundant gDMD EpiTYPER amplicons for the wild-type as well as mutant specimens at the respective time points, methylation was less in DNMT1o-deficient placental tissues at embryonic 12.5 day (E12.5), E15.5 and E17.5. At E12.5 it showed a significant reduction in the average methylation for all gDMDs (wild-type and mutant placentas) [53]. The average gDMD methylation was 0.283 for a total of 23 E15.5 DNMT1o-deficient placentas, which was significantly lower as compared to the wild-type 0.382 [54,55]. In one study, Meta-analysis was carried out in placenta by RNA microarray in 116 pre-eclamptic and 139 normotensive pregnancies using statistical and standard bioinformatics procedures [43], where pathway analysis of the expression signature in genes interactions were deduced as well as differentially expressed genes resulting to 388-gene meta-signatures of pre-eclamptic placenta. The analysis indicated the role of hypoxia/HIF1A pathway in the expression of pre-eclamptic gene profile which was consistence with previous reports [56]. Analysis of protein interaction networks showed that CREBBP/EP300 is a new element key to pre-eclamptic placental transcriptome and there is a high incidence of preeclampsia in pregnant women carrying fetus with a mutation in CREBBP/EP300 which normally lead to Rubinstein-Taybi Syndrome [57]. From the 388-gene-preeclampsia meta-signature reported, important information can be generated on the role of these genes in the placental tissue such as CREBBP/EP300 and the related pathways, which can be utilized as functional molecules or biomarkers in preeclampsia. Understanding the molecular basis of preeclampsia may help in development of therapeutic measures to alleviate placental pathologies [58].
Placental Gene Deletion
Achaete-Scute Family BHLH Transcription Factor 2 (Ascl2) gene deletion in the mouse Kcnq1 cluster results to fetal lethality because of the limited development of placenta labyrinth and the resulting in build-up of trophoblast giant cells (TGCs) at E10.5 [59]. Deletion of Cdkn1c or Phlda2 genes found within the Kcnq1 cluster, lead to placental overgrowth [60,61] as well as transgenic over-expression of Cdkn1c or Phlda2 that leads to impaired placental growth [62- 64]. The growth and development of the placenta also relies on Igf2, which is a significant part of H19 gene imprinting cluster; as a result, deletion of Igf2 leads to growth restriction of both the placenta and the fetus while Igf2 overexpression lead to overgrowth of the placenta as well as the fetus [65]. In addition, when other imprinted genes are deleted that are not located within the Kcnq1 or H19 clusters, there is also abnormalities in placental phenotypes as in the case of deletion of Igf2r, Growth-factor receptor bound protein 10 (Grb10), or Mesoderm-specific transcript (Mest) which may alter placental growth while deletion of Retrotransposon Gag Like 1 (Rtl1) or Paternally expressed 10 (Peg10) impairs labyrinth development [66]. Mitogen-activated protein kinase kinase 1(Map2k1) also plays an important role in myeloid translocation gene (MTG) formation and syncytiotrophoblast layer II cells (SynTII) differentiation.
Deletion of the two Map2k1 alleles in SynT-II in Gcm1Cre mouse line implies that they are not relevant for placenta formation since Map2k1flox/− Rosa+/lacZ Tg+/Gcm1Cre mice were found to be viable [67]. Placenta taken from Map2k1flox/− Rosa+/lacZ Tg+/Gcm1Cre mutants resembled the control group. X-Gal staining indicated that the activation of RosalacZ Cre reporter allele in SynT-II. Moreover, absence of Map2k1 functions in SynT-II result to formation of MTG but do not have an impact on the survival of the embryo [68]. Multilayers of trophoblast giant cells (TGCs) are seen at the maternal-fetal interface in Rac1d/d mice as opposed to a single or double layer seen in Rac1f/f mice. Spongiotrophoblast cells which are a subtype of TGCs are spatially distributed are seen from the expression of TPBPA biomarkers [69] is similar to placenta in Rac1d/d and Rac1f/f mice. in Rac1d/d mice progression of the pregnancy to day 12 showed a high disorganization of the placentae with malformed layers and labyrinth [70]. Decidual expression of Rac1 helps to regulate TGCs proliferation and differentiation at the maternal-fetal interface and allows for correct placenta formation and development [71].
Placenta Gene Mutations
Mutations such as E1A binding protein p300 (EP300) and CREBBP have been associated with disorders during placenta and fetal development such as Rubinstein-Taybi syndrome (RSTS, OMIM 180849 and 613684). RSTS is an autosomal dominant congenital disorder that affects one in every 100,000 newborns. The condition is characterized by facial dysmorphism, mental retardation, skeletal disorders and postnatal growth deficiency [72]. Many of these mutations are due de novo and could be as a result of extensive deletions or point mutations. Though there lacks a clear correlation between the genotype and the phenotype, lack of CREBBP/EP300 histone acetyltransferase action may result to RSTS [73]. A number of preeclampsia cases result to a birth of a child with RSTS [74]. As a result, the fetal genotype is implicated with RSTS and affects the placenta function which leads to high incidences of preeclampsia cases [75]. These findings can be incorporated in the meta-analysis data in non-RSTS mothers to indicate that low CEBBP/EP300 levels impair placenta functioning which increases the chances of the mother developing preeclampsia [76].
Conclusion
There are limited longitudinal studies on placental gene expression in human partly because of ethical constraints. However, studies in mice indicate that transition occur during midgestation and same cellular subsets may express different genes without necessarily causing changes in the placental morphology in humans, microarray data from basal plate biopsy from the maternal fetal interface indicate remarkable changes from mid-gestation to term. Plethora of studies on differential gene expression profile are available both for healthy and impaired placentas, particularly in preeclampsia. However, there are limited numbers of studies on differential gene expression profile during normal gestational development of placenta in human. Only a single microarray study has addressed this gap. This review aimed to explore the molecular pathways as well as physiological changes that occur in the course of gestation that may affect the placental structure and function. We therefore hypothesized that the molecular reorganization and phenotypic variations are needed for normal placental development and can be deduced from the level of genes expressed. More studies are therefore needed to ascertain this.
Acknowledgement
We are deeply grateful to Obstetrics and gynecology department of Tongji Hospital affiliated to Tongji Medical College of Huazhong University of Science and Technology for their overwhelming support and reading material. Special thanks to Michael who dedicated his time in editing this manuscript.
Funding
This study was funded by the research grants from the National Key Research and Development Program of China (2018YFC1002900) and the National Nature Science Foundation of China 41671497.
Conflict of Interest
The authors declare that they have no competing interests.
Author’s Contribution
All authors contributed equally to the work.
References
- Mikheev AM, Tomohiro Nabekura, Amal Kaddoumi, Theo K Bammler, Rajgopal Govindarajan, et al. (2008) Profiling gene expression in human placentae of different gestational ages: an OPRU Network and UW SCOR Study. Reprod Sci 15(9): 866-877.
- Chen HW, Jeremy J W Chen, Chii Ruey Tzeng, Han Ni Li, Shu Ju Chang, et al. (2002) Global analysis of differentially expressed genes in early gestational decidua and chorionic villi using a 9600 human cDNA microarray. Mol Hum Reprod 8(5): 475-484.
- Ng EKO, Nancy BY Tsui, Tze K Lau, Tse N Leung, Rossa WK Chiu, et al. (2003) mRNA of placental origin is readily detectable in maternal plasma. Proc Natl Acad Sci USA 100(8): 4748-4753.
- Wataganara T, Erik S Leshane, Angela Y Chen, Lisa M Sullivan, Inga Peter, et al. (2004) Circulating cell-free fetal nucleic acid analysis may be a novel marker of fetomaternal hemorrhage after elective first-trimester termination of pregnancy. Ann N Y Acad Sci 1022: 129-134.
- Tsui NBY, SSC Chim, RWK Chiu, TK Lau, EKO Ng, et al. (2004) Systematic micro-array based identification of placental mRNA in maternal plasma: towards non-invasive prenatal gene expression profiling. Journal of medical genetics 41(6): 461-467.
- Ng EKO, Tse N Leung, Nancy B Y Tsui, Tze K Lau, Nirmal S Panesar, et al. (2003) The concentration of circulating corticotropin-releasing hormone mRNA in maternal plasma is increased in preeclampsia. Clin Chem 49(5): 727-731.
- Roberts RM, Green JA, Schulz LC (2016) The evolution of the placenta. Reproduction 152(5): R179-R189.
- Korneeva KL, RR Rodriguez, SV Ralchenko, OV Martunovska, AO Frolova, et al. (2016) Expression of genes, Encoding the enzymes of cysteine metabolism in human placenta in the first and third trimesters of uncomplicated pregnancy. Ukr Biochem J 88(1): 88-98.
- Sitras V, Fenton C, Paulssen R, Vårtun Å, Acharya G (2012) Differences in Gene Expression between First and Third Trimester Human Placenta: A Microarray Study. PLoS One 7(3): 1-7.
- Winn VD, Ronit Haimov Kochman, Agnes C Paquet, Y Jean Yang, MS Madhusudhan, et al. (2007) Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology 148(3): 1059-1079.
- Gheorghe CP, Mohan S, Oberg KC, Longo LD (2007) Gene expression patterns in the hypoxic murine placenta: A role in epigenesis? Reprod Sci 14(3): 223-233.
- Small HY, Christine Akehurst, Liliya Sharafetdinova, Martin W McBride, John D McClure, et al. (2017) HLA gene expression is altered in whole blood and placenta from women who later developed preeclampsia. Physiol Genomics 49(3): 193-200.
- Enquobahrie DA, Margaret Meller, Kenneth Rice, Bruce M Psaty, David S Siscovick, et al. (2008) Differential placental gene expression in preeclampsia. Am J Obstet Gynecol 199(5): 566.e1-e11.
- Tsai S, NE Hardison, AH James, AA Motsinger Reif, SR Bischoff, et al. (2011) Transcriptional profiling of human placentas from pregnancies complicated by preeclampsia reveals disregulation of sialic acid acetylesterase and immune signalling pathways. Placenta 32(2): 175-182.
- Goldman Wohl D, T Cesla, Y Smith, C Greenfield, R Dechend, et al. (2013) Expression profiling of autophagy associated genes in placentas of preeclampsia. Placenta 34(10): 959-962.
- Brew O, Sullivan MHF, Woodman A (2016) Comparison of normal and pre-eclamptic placental gene expression: A systematic review with meta-analysis. PLoS One.
- Yokomine T, Hisao Shirohzu, Wahyu Purbowasito, Atsushi Toyoda, Hisakazu Iwama, et al. (2005) Structural and functional analysis of a 0.5-Mb chicken region orthologous to the imprinted mammalian Ascl2/Mash2-Igf2-H19 region. Genome Res 15(1): 154-165.
- Hemberger M (2007) Epigenetic landscape required for placental development. Cell Mol Life Sci 64: 2422-2436.
- Novakovic B, V Rakyan, HK Ng, U Manuelpillai, C Dewi, et al. (2008) Specific tumour-associated methylation in normal human term placenta and first-trimester cytotrophoblasts. Mol Hum Reprod 14(9): 547-554.
- Sun M, J Kingdom, D Baczyk, SJ Lye, SG Matthews, et al. (2006) Expression of the multidrug resistance P-glycoprotein, (ABCB1 glycoprotein) in the human placenta decreases with advancing gestation. Placenta 27(6-7): 602-609.
- Gil S, Saura R, Forestier F, Farinotti R (2005) P-glycoprotein expression of the human placenta during pregnancyPlacenta 26(2-3): 268-270.
- Yeboah D, M Sun, J Kingdom, D Baczyk, SJ Lye, et al. (2006) Expression of breast cancer resistance protein (BCRP/ABCG2) in human placenta throughout gestation and at term before and after labor. Can J Physiol Pharmacol 84(12): 1251-1258.
- Mathias AA, Hitti J, Unadkat JD (2005) P-glycoprotein and breast cancer resistance protein expression in human placentae of various gestational ages. Am J Physiol Regul Integr Comp Physiol 289(4): R963-R969.
- Meyer zu Schwabedissen HE, Annette Dreisbach, Elke Hammer, Christoph Fusch, Michael Hecker, et al. (2006) Direct mass spectrometric identification of ABCB1 (P-glycoprotein/MDR1) from the apical membrane fraction of human placenta using fourier transform ion cyclotron mass spectrometry. Pharmacogenet Genomics 16(6): 385-389.
- Polgar O, Robey RW, Bates SE (2008) ABCG2: structure, function and role in drug response. Expert Opin Drug MetabToxicol 4(1): 1-15.
- Chang X (2007) A molecular understanding of ATP-dependent solute transport by multidrug resistance-associated protein MRP1. Cancer Metastasis Rev 26(1): 15-37.
- Nagashige M, F Ushigome N Koyabu, K Hirata, M Kawabuchi, et al. (2003) Basal membrane localization of MRP1 in human placental trophoblast. Placenta 24(10): 951-958.
- Serrano MA, RIR Macias, O Briz, MJ Monte, AG Blazquez, et al. (2007) Expression in human trophoblast and choriocarcinoma cell lines, BeWo, Jeg-3 and JAr of genes involved in the hepatobiliary-like excretory function of the placenta. Placenta 28(2-3): 107-117.
- Oros D, M Strunk, P Breton, C Paules, R Benito, et al. (2017) Altered gene expression in human placenta after suspected preterm labour. Placenta 55: 21-28.
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402-408.
- Proudhon C, Bourc’his D (2010) Identification and resolution of artifacts in the interpretation of imprinted gene expression. Brief Funct Genomics 9: 374-384.
- Fitzpatrick GV, Soloway PD, Higgins MJ (2002) Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet 32(3): 426-431.
- Wutz A, OW Smrzka, N Schweifer, K Schellander, EF Wagner, et al. (1997) Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389(6652): 745-749.
- Sleutels F, Zwart R, Barlow DP (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415(6873): 810-813.
- Nagano T, Jennifer A Mitchell, Lionel A Sanz, Florian M Pauler, Anne C Ferguson Smith, et al. (2008) The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322(5908): 1717-1720.
- Regha K, Mathew A Sloane, Ru Huang, Florian M Pauler, Katarzyna E Warczok, et al. (2007) Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell 27(3): 353-366.
- Paradowska E, Blach Olszewska Z, Gejdel E (1997) Constitutive and induced cytokine production by human placenta and amniotic membrane at term. Placenta 18(5-6): 441-446.
- Pandey RR, Tanmoy Mondal, Faizaan Mohammad, Stefan Enroth, Lisa Redrup, et al. (2008) Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-Specific Transcriptional Silencing through Chromatin-Level Regulation. Mol Cell 32(2): 232-246.
- Monk D (2015)Genomic imprinting in the human placenta. Am J Obstet Gynecol 213(4): S152-S162.
- Rahat B, Mahajan A, Bagga R, Hamid A, Kaur J (2017) Epigenetic modifications at DMRs of placental genes are subjected to variations in normal gestation, pathological conditions and folate supplementation. Sci Rep 7: 40774.
- Frost JM, Moore GE (2010) The Importance of Imprinting in the Human Placenta. PLOS Genet 6: 1-9.
- Maulik D, Alok De, Louis Ragolia, Jodi Evans, Dmitry Grigoryev, et al. (2016) Down-regulation of placental neuropilin-1 in fetal growth restriction. Am J Obstet Gynecol 214(2): 279.e1-279.e9.
- van Uitert M, Perry D Moerland, Daniel A Enquobahrie, Hannele Laivuori, Joris A M van der Post, et al. (2015) Meta-Analysis of Placental Transcriptome Data Identifies a Novel Molecular Pathway Related to Preeclampsia. PLoS One 10: e0132468-e0132468.
- Yang Z, Yongbin Chen, Yu Fu, Yihao Yang, Ya Zhang, et al. (2014) Meta-analysis of differentially expressed genes in osteosarcoma based on gene expression data. BMC Med Genet 15: 80.
- Kleinrouweler CE, Miranda van Uitert, Perry D Moerland, Carrie Ris Stalpers, Joris A M van der Post, et al. (2013) Differentially expressed genes in the pre-eclamptic placenta: a systematic review and meta-analysis. PLoS One 8(7): e68991-e68991.
- Ito Y, Daitoku H, Fukamizu A (2009) Foxo1 increases pro-inflammatory gene expression by inducing C/EBPbeta in TNF-alpha-treated adipocytes. Biochem Biophys Res Commun 378(2): 290-295.
- Acosta AM, Escalona M, Maiz A, Pollak F, Leighton F (2002) Determination of the insulin resistance index by the Homeostasis Model Assessment in a population of Metropolitan Region in Chile. Rev Med Chil 130(11): 1227-1231.
- Wagner PK, Otomo A, Christians JK (2011) Regulation of pregnancy-associated plasma protein A2 (PAPPA2) in a human placental trophoblast cell line (BeWo). Reprod Biol Endocrinol 9: 48.
- Wagner P, Wagner P (2011) Pregnancy-associated plasma protein a2 by thesis submitted in partial fulfillment of the requirements for the degree of In the Department of Biological Sciences. 2.
- Wang J, Qing Qiu, Maliha Haider, Michael Bell, Andrée Gruslin, et al. (2009) Expression of pregnancy-associated plasma protein A2 during pregnancy in human and mouse. The Journal of endocrinology 202(3): 337-345.
- (2011) Manuscript, ANIH Public Access 80: 1754-1761.
- Robinson WP, Price EM (2015) The human placental methylom. Cold Spring Harb Perspect Med 5(5): a023044.
- Koppes E, Himes KP, Chaillet JR (2015) Partial loss of genomic imprinting reveals important roles for Kcnq1 and Peg10 imprinted domains in placental development. PLoS One 10(8): e0135202.
- Deshpande SS, Balasinor NH (2018) Placental Defects: An Epigenetic Perspective. Reprod Sci 25(8): 1143-1160.
- Takahashi Y, K Mitani, K Kuwabara, T Hayashi, M Niwa, et al. (1994) Methylation imprinting was observed of mouse mo-2 macrosatellite on the pseudoautosomal region but not on chromosome 9. Chromosoma 103(7): 450-458.
- Nishizawa H, K Pryor Koishi, T Kato, H Kowa, H Kurahashi, et al. (2007) Microarray analysis of differentially expressed fetal genes in placental tissue derived from early and late onset severe pre-eclampsia. Placenta 28(5-6): 487-497.
- Conditions G Genetics Home Reference Rubinstein-Taybi syndrome.
- Fergelot P, Martine Van Belzen, Julien Van Gils, Alexandra Afenjar, Christine M Armour,et al. (2016) Phenotype and Genotype in 52 Patients with Rubinstein-Taybi Syndrome Caused by EP300 Mutations. American Journal of Medical Genetics Part A 170(12): 3069-3082.
- Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL (1994) Essential role of Mash-2 in extraembryonic development. Nature 371(6495): 333-336.
- Frank D, Weiwei Fortino, Lorraine Clark, Raymond Musalo, Wenxian Wang, et al. (2002) Placental overgrowth in mice lacking the imprinted gene Ipl. Proc Natl Acad Sci USA 99(11): 7490-7495.
- Tunster SJ, Van de Pette M, John RM (2011) Fetal overgrowth in the Cdkn1c mouse model of Beckwith-Wiedemann syndrome. Dis Model Mech 4(6): 814-821.
- Salas M, Rosalind John, Anjana Saxena, Sheila Barton, Dale Frank, et al. (2004) Placental growth retardation due to loss of imprinting of Phlda2. Mech Dev 121(10): 1199-1210.
- Tunster SJ, Tycko B, John RM (2010) The imprinted Phlda2 gene regulates extraembryonic energy stores. Mol Cell Biol 30(1): 295-306.
- Andrews SC, Michelle D Wood, Simon J Tunster, Sheila C Barton, M Azim Surani, et al. (2007) Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7. BMC Dev Biol 7: 53.
- Sun FL, Dean WL, Kelsey G, Allen ND, Reik W (1997) Transactivation of Igf2 in a mouse model of Beckwith-Wiedemann syndrome. Nature 389(6653): 809-815.
- Charalambous M, Michael Cowley, Fleur Geoghegan, Florentia M Smith, Elizabeth J Radford, et al. (2010) Maternally-inherited Grb10 reduces placental size and efficiency. Dev Biol 337(1): 1-8.
- Nadeau V, Stéphanie Guillemette, Louis François Bélanger, Olivier Jacob, Sophie Roy, et al. (2009) Map2k1 and Map2k2 genes contribute to the normal development of syncytiotropho blasts during placentation. Development 136(8): 1363-1374.
- Nadeau V, Charron J (2014) Essential role of the ERK/MAPK pathway in blood-placental barrier formation. Development 141(14): 2825-2837.
- Hu D, Cross JC (2010) Development and function of trophoblast giant cells in the rodent placenta. Int J Dev Biol 54(2-3): 341-354.
- Rossant J, Cross JC (2001) Placental development: lessons from mouse mutants. Nat Rev Genet 2(7): 538-548.
- Cross JC (2005) How to make a placenta: mechanisms of trophoblast cell differentiation in mice--a review. Placenta 26: S3-S9.
- van Belzen M, Bartsch O, Lacombe D, Peters DJM, Hennekam RCM (2011) Rubinstein-Taybi syndrome (CREBBP, EP300). Eur J Hum Genet 19: 118-120.
- Roelfsema JH, Peters DJM (2007) Rubinstein-Taybi syndrome: clinical and molecular overview. Expert Rev Mol Med 9(23): 1-16.
- Negri G, D Milani, P Colapietro, F Forzano, M Della Monica, et al. (2015) Clinical and molecular characterization of Rubinstein-Taybi syndrome patients carrying distinct novel mutations of the EP300 gene. Clin Genet 87(2): 148-154.
- Tsai ACH, Cherilyn J Dossett, Carol S Walton, Andrea E Cramer, Patti A Eng, et al. (2011) Exon deletions of the EP300 and CREBBP genes in two children with Rubinstein-Taybi syndrome detected by aCGH. Eur J Hum Genet 19(1): 43-49.
- Wieczorek D, Oliver Bartsch, Stanislav Lechno, Jürgen Kohlhase, Dorien J M Peters, et al. (2009) Two adults with Rubinstein-Taybi syndrome with mild mental retardation, glaucoma, normal growth and skull circumference, and camptodactyly of third fingers. Am J Med Genet A 149A(12): 2849-2854.

 Review Article
Review Article