Abstract
Background: Neuroblastoma is the highest mortality rate extracranial soild tumor in childhood. Accumulating evidence indicated that long noncoding RNAs (lncRNAs) are widely expressed in neuroblastoma, and playing an important role in the development and progression.
Methods: RNA sequencing was conducted to identify differentially expressed lncRNAs in four Ⅲ phase and four Ⅳ phase tumor tissues of neuroblastoma. RT-qPCR was carried out to validate the result of sequencing. Clinical information was reviewed to analyze the relationship between lncRNA and clinical characteristics. The public database R2 was used to analyze prognosis.
Result: Differentially expressed lncRNAs were identified. LRRC75A-AS1 was the overexpressed lncRNA in Ⅳ phase patients. RT-qPCR was conducted in tumor tissues, confirming the tendency with sequencing. And higher expression of LRRC75A-AS1 was associated with N-MYC (p < 0.001), advanced stage (p = 0.029), Risk group (p = 0.027). Furthermore, LRRC75A-AS1 was correlated with Shimada classification(p = 0.046), LDH level (r = 0.390, p = 0.003), D-Dimer level (r = 0.338, p = 0.012) , and NSE level (r = 0.284, p = 0.05). The neuroblastoma dataset shows that patients with overexpressed LRRC75A-AS1 have a worse prognosis than down-expressed.
Conclusion: LRRC75A-AS1 is associated with clinical characteristics of neuroblastoma and may function as a prognostic predictor or a therapeutic target.
Keywords: Biomarker; lncRNA LRRC75A- AS1; Neuroblastoma
Abbreviations: NB: Neuroblastoma; lncRNAs: Long Noncoding RNAs; LDH: Lactate Dehydrogenase; NSE: Neuron-Specific Enolase; ORFs: Open Reading Frames; uFH: Unfavorable Histology; FH: Favorable Histology; NA: Not Amplified; NM: Not Metastasis; PD: Progressive Disease; VMA: Vanilla Mandelic; SNHG29: Small Nucleolar RNA Host Gene 29; TJ: Tight Junction; CRC: Colorectal Carcinoma
Introduction
Neuroblastoma (NB) is a sympathetic embryonic tumor
originating from the neural crest of the embryonic sympathetic
nervous system,it is the most common extracranial solid tumor
in children which accounts for 7-10% of all childhood cancer
mortality [1-5]. In order to take tailored treatment approaches
for neuroblastoma, pediatric cooperative groups introduce risk
factors including clinical stage, age, histologic category, grade of
tumor differentiation, MYCN status, DNA ploidy, and 11q exception
[6,7]. High-risk neuroblastoma patients often have unfavorable
outcomes, with the 5-year overall survival rate less than 50% [6].
The application of genetic difference analysis promote accurate
stratification has attracted widespread attention. Therefore, it is of
great practical significance and theoretical value to explore effective
drug targets and better biomarkers for advanced neuroblastoma.
Long noncoding RNAs (lncRNAs) refer to endogenous RNAs that
are longer than 200 nucleotides and lack of specific complete open
reading frames (ORFs) and the function of protein-coding [8,9].
Thus they were once considered a part of transcriptional noise,
but now have been proved as potential key regulators of promoting
or maintaining tumorigenesis and the development of cancer,
having clinical potential as prognostic biomarkers for targeted
therapeutics and interventions in various cancers[4,10]. Several
lines of evidence have shown that lncRNAs have been implicated
in initiation and progression of neuroblastoma [5,6], and lncRNAbased
prognostic biomarkers have been proposed for tumor
stratification and predicting survival. Therefore, deep investigating
of the roles and mechanisms of lncRNAs in tumorigenesis provides
promises in developing new biomarkers and molecular-targeted
therapy. Our research aims to identify lncRNA-based biomarkers that could be used for prognosis prediction and treatment. The
purpose of this study was to explore the relationship between
lncRNAs and clinicopathological parameters in neuroblastoma
patients, to further explore the lncRNAs that lead to the invasion
and metastasis of neuroblastoma. In the first, we conducted RNAsequencing
to identify differentially expressed lncRNAs in 4 Ⅲ stage
and 4 Ⅳ stage patients’ tumor tissues of neuroblastoma, and we
identified a lncRNA named LRRC75A-AS1 was upregulated lncRNAs
in Ⅳ stage neuroblastomas, Futher to explore the relationship
between LRRC75A-AS1 and clinical characteristics ,RT-qPCR was
carried out to detect 57 cases of neuroblastoma.
Materials and Methods
Patient
A total of 57 cases of fresh primary tumor tissues with pathologically diagnosed neuroblastoma were collected in this study approved by the ethics committee of the Children’s Hospital of Chongqing Medical University from August 2014 to May 2019. And subjects (or their parents or guardians) have given their written informed consent.
The Inclusion Criteria
a) Pathologically diagnosed NB.
b) Primary tumor without any treatment.
c) Written informed consent was obtained from the
guardians.
The Exclusion Criteria
a) History of other malignant disease.
b) Recurrent or treated disease.
c) The quality of tissues was unqualified.
Clinical features of these patients at diagnosis including age, gender, tumor size, INSS stage, risk group, MYCN status, tumor biomarkers, and metastasis were retrospectively collected. All fresh tissue specimens were preserved in −80℃ until use. R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl) was used to investigate the relationship between lncRNA expression and overall survival with neuroblastoma patients.
Expression Profile Analysis of RAN-Sequencing
The RAN-sequencing was employed to identify neuroblastomarelated RNAs. After hybridization and washing with samples, 8 samples of extracted RNA of neuroblastoma tumor tissues (4 III stage neuroblastoma and 4 IV stage neuroblastoma) were analyzed.
RNA Extraction and Real Time qRT-PCR Analysis
Total RNA of specimens was extracted using RNA extraction reagent kit (Bio Teke). RNA concentration and purity were measured by NonoDrop (Thermo Scientific). cDNA was reverse transcribed with the Prime Script RT reagent Kit ((Takara Biotechnology Co., Ltd, China) from 1000 ng of total RNA. The real-time qPCR analyses were carried out using SYBR GREEN Premix ExTaq kit (Danfeng, China) by CFX96 Cycler System. Relative RNA expression was computed by 2-ΔΔCt method with normalization to human β-actin. The primers for LRRC75A-AS1 are: F 5′- AGCTCACAGCACACCTGGCTA-3′and R 5′-AGCTGAGGCAGGAGGACCAT-3′, and the primers for β-actin are: 5′-CCTGGCACCCAGCACAAT-3′ and R 5′-GGGCCGGACTCGTCATAC-3′.
Statistical Analysis
Statistical analyses were conducted by SPSS 23.0 (IBM Corporation. Armonk, NY, USA). Graphical depiction of data was generated by GraphPad Prism.v5.0. (GraphPad Software, Inc., La Jolla, CA). In the statistical analysis, a two-sided p value <= 0.05 was considered statistically significant. Differentially expressed lncRNAs were identified through fold change as well as P value calculated with t-test. The threshold set for up- and downregulated genes was a fold change > = 2.0 and a p value <= 0.05. For qualitative data, the χ2 test or Fisher exact test was used to evaluate the significance between groups. For quantitative data, Kruskal-Wallis test was used to analyze the significance between individual groups. The correlations were analyzed by Spearman correlation analysis. The prognostic relationship was evaluated using Kaplan-Meier.
Result
Screening for Differentially Expressed lncRNAs
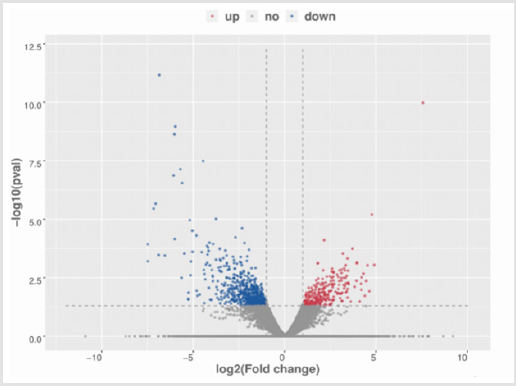
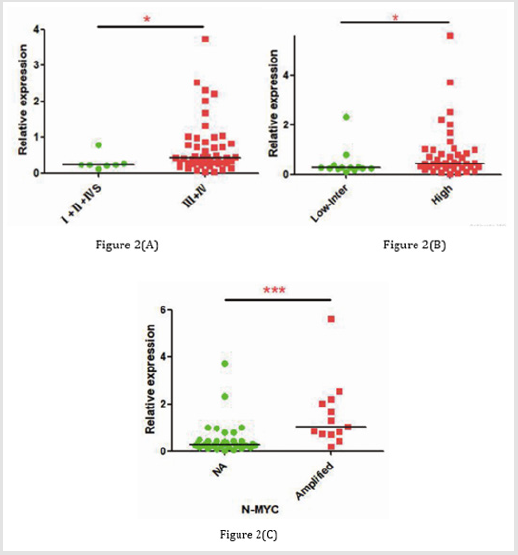
To investigate marrow metastasis-related RNA expression profile in neuroblastoma tissues, we analyzed 8 tissue samples of neuroblastoma (4 III stage and 4 IV stage patients) by using the RNA-sequencing. The pathological characteristics of the 8 patients are listed in Table 1. With the threshold set for up- and downregulated genes of a fold change >= 2.0 and a p value <= 0.05, 1043differentially expressed lncRNAs were identified between III and IV stage tumor samples, including 458 upregulated lncRNAs and 585 downregulated lncRNAs. Among them, we found that LRRC75A-AS1 was the overexpressed lncRNA in IV stage patients with the fold change of 3.19 (p = 0.02, (Figure 1). To determine the tendency of sequence, RT-qPCR was carried out to measure LRRC75A-AS1expression level of 57 neuroblastoma tumor tissues, including the 8 patients ‘tumor tissues for sequencing. The relative expression of LRRC75A-AS1 ranged from 0.03 to 5.62, with the median value of 0.39. Compared to the Ⅲ stage tissues, expression of LRRC75A-AS1was higher in Ⅳ stage tissues, however it’s not statistically significant (p =0.152). But compared to the early stage tissues, expression of LRRC75A-AS1was higher in advanced stage tissues (p =0.029) (Figure 2A). LRRC75A-AS1 was higher in highrisk than intermediate-risk and low-risk neuroblastoma (p = 0.027) (Figure 2B). LRRC75A-AS1 was higher in N-MYC amplified disease than N-MYC Not-amplified disease (p < 0.001) (Figure 2C). The results suggesting that lncRNA LRRC75A-AS1 may play a significant role in the pathogenesis and development of neuroblastoma.
Figure 1: The volcano plot showed differentially expressed lncRNAs between Ⅲ stage and Ⅳ stage neuroblastoma tumor samples.
Figure 2: The correlation between expression of LRRC75A-AS1 and Stage, Risk classification, N-MYC.
(A) Relative expression of LRRC75A-AS1 in early (I, II, IVs) and advanced (III, IV) stage disease.
(B) Relative expression of LRRC75A-AS1 in low and intermediate (low + inter) and high-risk neuroblastoma.
(C) Relative expression of LRRC75A-AS1 in N-MYC(MYCN) not amplified, and N-MYC amplified.
Table 1: Clinicopathological parameters of 8 patients whose tumors were used for RNA-sequencing.
Note: INSS: International Neuroblastoma Staging System; Inter: Intermediate; uFH: Unfavorable Histology; NA: Not Amplified; NM: Not Metastasis; PD: Progressive Disease
Correlations Between the Expression Level of LRRC75AAS1 and Clinical Characteristics
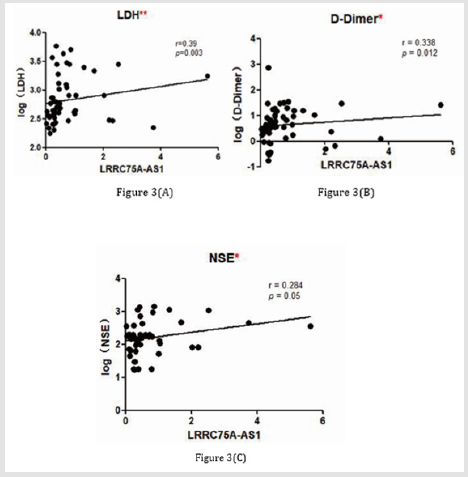
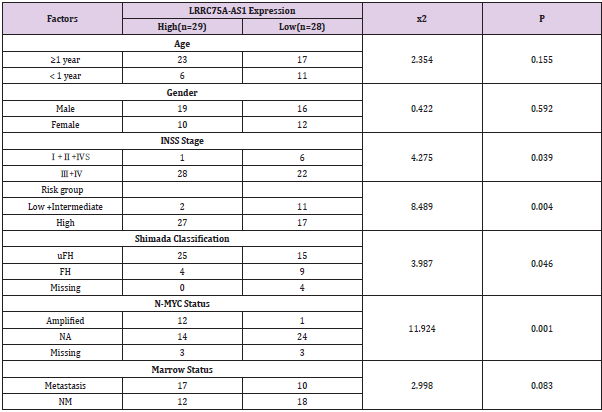
The 57 neuroblastoma patients were divided into two groups (high or low) based on the median value of LRRC75A-AS1 expression level (Table 2). We found that high LRRC75A-AS1 expression level in tumor tissues was associated with advanced INSS stage (p = 0.039), risk group (p = 0.004), N-MYC status (p = 0.001), shimada classification (p = 0.046). Moreover, we identified that LRRC75AAS1 expression was correlated with serum lactate dehydrogenase (LDH) level (r = 0.390, p = 0.003) (Figure 3A), D-Dimer level (r = 0.338, p = 0.012) (Figure 3B), and serum neuron-specific enolase (NSE) level (r = 0.284, p = 0.05) (Figure 3C) ,but has no correlation with ki-67 level (p = 0.163) ,Vanilla mandelic acid(VMA) level (p = 0.073) ,and tumor size (p = 0.515).
Table 2: Relationships between the expression of LRRC75A-AS1 and clinicopathological parameters in neuroblastoma
Note: INSS: International Neuroblastoma Staging System; uFH: Unfavorable Histology; FH: Favorable Histology; NA: Not Amplified; NM: Not Metastasis
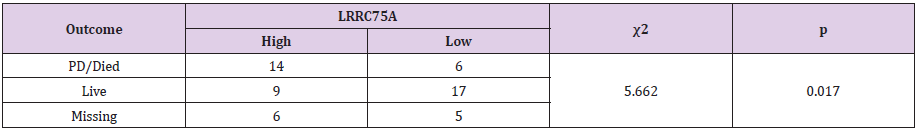
Relations Between Expression Level of LRRC75A-AS1 and Prognosis in Neuroblastoma Patients
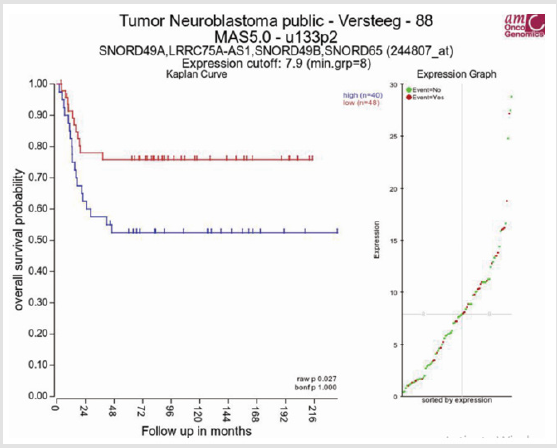
Owing to the small sample size and limited follow-up time of partial patients, we cannot analyze the correlation between overall survival and LRRC75A-AS1 in our patients. Thence, we see progressive disease or died as outcome indicator to investigate the relationship of LRRC75A-AS1 expression level and prognosis with neuroblastoma patients. The results show that high expression of LRRC75A-AS1 was correlated with poor prognosis (χ2=5.662, p = 0.017) (Table 3). Further to investigate the relationship of LRRC75A-AS1 expression and overall survival in neuroblastoma patients. R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl) was used to analyze the relationship (Figure 4). Kaplan–Meier analysis demonstrated elevated LRRC75A-AS1 expression levels were associated with poor prognosis, whereas low expression of LRRC75A-AS1 was associated with favorable outcome in the Versteeg dataset consisting of a cohort of 88 neuroblastoma patients (n = 88, log-rank: p = 0.027). To sum up, our analysis of LRRC75A-AS1 with clinical features and microarray dataset indicated that LRRC75A-AS1 was a novel prognostic marker in neuroblastoma.
Figure 4: The relationship of LRRC75A-AS1 expression and overall survival in neuroblastoma patients. Kaplan-Meier survival plot was downloaded from R2 Genomics Analysis and Visualization Platform (+).
Discussion
Over the last decade, with the extensive development of genomic
transcription study, accumulative studies indicated that lncRNA
have risen to prominence with important roles in a broad range
of biological processes. Recent studies have reported that several
lncRNAs, for example, MALATA, CASC15, LOC440896, LINC00632,
IGF2-AS have been implicated in characteristics and prognosis
of neuroblastoma [5,6,11]. Therefore, lncRNA have the potential
to serve as novel biomarkers for neuroblastoma diagnosis or
prognosis. However, the biological functions of most lncRNAs have
yet to be explored. High-risk neuroblastoma patients often have
unfavorable outcomes, it is one of the biggest obstacles to improve
overall survival of neuroblastoma. Hence, it is urgent to investigate
the novel genes and illustrate the molecular mechanisms of
neuroblastoma. The main purpose of this study was to investigate
the differential expression of LRRC75A-AS1 in neuroblastoma, and
to find new prognostic and diagnostic markers for neuroblastoma.
The results of this study show that LRRC75A-AS1 is up-regulated in
neuroblastoma.
Small nucleolar RNA host gene 29 (SNHG29) was also known
as LRRC75A-AS1, TSAP19, C17orf45, NCRNA00188, FAM211AAS1,
C17or f76-AS1, it’s a long noncoding RNA that leucine rich
repeat containing 75 A-antisense RNA1 is located on 17p11.2 [12].
Emerging reports has revealed that LRRC75A-AS1 was involved
in several biological processes through modulation of signaling
pathway, Jeong et al. [13] reported that LRRC75A-AS1 can regulate
the vascular calcification negatively, and might act as a possible
target in the treatment of vascular calcification. Wang et al. [14]
found that LRRC75A-AS1 can regulate the expression of tight
junction (TJ) proteins through LRRC75A, affecting the inflammatory
responses of bovine mammary epithelial cells. Leavey K et al. [15]
show that LRRC75A is abnormally expressed in the process of
normal villous maturation.
In cancers, LRRC75A-AS1 has been served as a crucial regulator
in a variety types of cancers including osteosarcoma [12,16],
colorectal carcinoma [17], breast cancer [18], gastric cancer [19],
glioblastoma [20] and acute myeloid leukemia [21]. Joeri Both et
al. [12,15] claimed that LRRC75A serves as malignant facilitatorin
in osteosarcoma. Jianxiong Chen et al. [16] reported that LRRC75AAS1
inhibits cell proliferation and migration in colorectal
carcinoma, and it might serve as an anti-oncogene for colorectal
carcinoma (CRC) tumorigenesis and advancement. Lizhang Han et
al. [20] found that SNHG2(LRRC75A-AS1) can regulates miR-223-
3p/CTNND1 axis to promote glioblastoma progression via Wnt/β-
catenin signaling pathway. FANGCE WANG et al. [21] proved that
LRRC75A-AS1 can significantly predict prognosis of acute myeloid
leukemia. These papers showed that LRRC75A-AS1 may become
a novel molecular marker for diagnosis and treatment of cancer.
However, little information of the prognostic value and the role of
LRRC75A-AS1 in neuroblastoma has been reported.
In the present study, we compare the expression profile of
lncRNA between 4 Ⅲ phase and 4 Ⅳ phase tumor tissues of
neuroblastoma by using the RNA-sequencing. We found that
LRRC75A-AS1 was the overexpressed lncRNA in IV stage patients
with the fold change of 3.19. To further confirm the relationship
of LRRC75A-AS1 in neuroblastoma, the RT-qPCR analysis was used to analyze the clinical tissue from neuroblastoma patients. These
experimental results showed that LRRC75A-AS1was obviously
high in advanced stage neuroblastoma, and high expression level
of LRRC75A-AS1 was associated with advanced stage disease, high
risk group, N-MYC Amplified, unfavorable histology, and the level of
LDH, D-Dimer and NSE, which are strong predictors for prognosis
of neuroblastoma. Furthermore, we used public neuroblastoma
dataset in R2 validated that overexpression of LRRC75A-AS1 was
correlated with unfavorable prognosis in neuroblastoma. But, the
expression of LRRC75A-AS1 was not correlated with ki-67 level,
Vanilla mandelic acid(VMA) level and tumor size, the inconsistency
may be caused by the small sample size of this study which did not
permit attainment of statistical significance.
The limitation of current experiments is the small sample size
resulting in limited statistical power, hence, it is still necessary
to expand the clinical sample size and patients should also be
long-term followed up to validate the prognosis in the public
neuroblastoma dataset in our cohort. This study merely proved
the relationship between LRRC75A-AS1 and tumor in clinic, so
more relevant basic experiments should be practiced on animals
and cells. Further studies are needed to illuminate the underlying
molecular mechanisms that LRRC75A-AS1 might promote the
tumorigenesis and progression of NB, as well as screening for
potential therapeutic target for neuroblastoma.
Conclusion
In conclusion, our study demonstrated that LRRC75A-AS1 was up-regulated in Ⅳ phase tumor. Further experiments revealed that overexpression of LRRC75A-AS1 in tumor tissues was associated with aggressive disease including INSS III, IV stage, high risk group, N-MYC amplified, uFH classification, high level of LDH, D-Dimer and NSE, and unfavorable overall survival. Thus LRRC75A-AS1 may function as a potential prognostic biomarker in neuroblastoma, and we conjecture a novel prognostic model including LRRC75AAS1 may predict the outcomes of neuroblastoma patient in clinical practice more accurately.
Statements
Acknowledgement
We are grateful for the help from all participators. This work was supported by the General project of clinical medicine research of Children’s Hospital Affiliated to Chongqing Medical University (No. YBXM-2019-003) and Chongqing Science and Technology Bureau Project (No. cstc2019jscx-msxmX0220 and cstc2016shmsztzx0042)
Statement of Ethics
This research complies with the guidelines for human studies. This study approved by the ethics committee of the Children’s Hospital of Chongqing Medical University and subjects (or their parents or guardians) have given their written informed consent.
Disclosure Statement
The authors have no conflicts of interest to declare
Funding Sources
This work was supported by grants from the General project of clinical medicine research of Children’s Hospital Affiliated to Chongqing Medical University (No.YBXM-2019-003) received by W.S., Chongqing Science and Technology Bureau Project (No. cstc2019jscx-msxmX0220 and cstc2016shms-ztzx0042) received by W.S. No funding bodies had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
WZ and WS designed the experiments, analyzed the data and revised the manuscript. WZ wrote the manuscript. WZ performed most of the experiments. LC, YC, PL and SJ collected tumor tissues and performed the experiments. All of the authors discussed the results and reviewed the manuscript.
References
- Saunders WB (2012) Seminars in pediatric surgery [J] 21(1): 2-14.
- Maris JM (2010) Recent Advances in Neuroblastoma[J]. N Engl J Med 362(23): 2202-2211.
- Westermann F, Schwab M (2002) Genetic parameters of neuroblastomas[J]. Cancer Letters 184(2):127-147.
- Lin C, Yang L (2018) Long noncoding RNA in cancer: wiring signaling circuitry[J]. Trends Cell Biol 28: 287-301.
- Irwin MS, Park JR (2015) Neuroblastoma Paradigm for Precision Medicine[J]. Pediatric Clinics of North America 62(1): 225-256.
- Pinto NR, Applebaum MA, Volchenboum SL, Katherine K Matthay, Wendy B London, et al. (2015) Advances in risk classifification and treatment strategies for neuroblastoma[J]. Clin Oncol 33(27): 3008-3017.
- Maris JM (2010) Recent advances in neuroblastoma[J]. N Engl J Med 362(23): 2202-2211.
- Tsai MC, Spitale RC, Chang HY (2011) Long intergenic noncoding RNAs: new links in cancer progression[J]. Cancer Res 71: 3-7.
- Deniz E, Erman B (2017) Long noncoding RNA (lncRNA), a new paradigm in gene expression contro[J]l. Funct Integr Genomics 17: 135-143.
- Arnes L, Liu Z, Wang J, Carlo Maurer, Irina Sagalovskiy, et al. (2019) Comprehensive characterisation of compartment-specific long non-coding RNAs associated with pancreatic ductal adenocarcinoma[J]. Gut: gutjnl-2017-314353.
- Srinivasulu, Yerukala, Sathipati, Yenching Lin, Shinn-Ying Ho (2019) Identification and characterization of the lncRNA signature associated with overall survival in patients with neuroblastoma[J]. Scientific reports 9(1): 5125.
- Both J, Wu T, Ten Asbroek AL, Baas F, Hulsebos TJ (2016) Oncogenic properties of candidate oncogenes in chromosome region 17p11.2p12 in human osteosarcoma[J]. Cytogenet Genome Res 150: 52-59.
- Jeong G, Kwon DH, Shin S, Choe N, Ryu J, et al. (2019) Long noncoding RNAs in vascular smooth muscle cells regulate vascular calcifification[J]. Sci Rep 9: 5848.
- Wang X, Wang H, Ming-Qing Gao (2020) LRRC75A antisense lncRNA1 knockout attenuates inflammatory responses of bovine mammary epithelial cells[J]. Int J Biol Sci 16(2): 251-263.
- Leavey K, Benton SJ, Grynspan D, Bainbridge SA, Morgen EK, et al. (2017) Gene markers of normal villous maturation and their expression in placentas with maturational pathology[J]. Placenta 58: 52-59.
- Both J, Wu T, Bras J, Gerard R. Schaap, Frank Baas, et al. (2012) Identification of Novel Candidate Oncogenes in Chromosome Region 17p11.2-p12 in Human Osteosarcoma[J]. PLOS ONE 7(1): e30907.
- Chen J, Lan J, Zhiwei Ye, ShiyuDuan, Yukun Hu, et al. (2019) Long noncoding RNA LRRC75A-AS1 inhibits cell proliferation and migration in colorectal carcinoma[J]. Exp Biol Med (Maywood) 244(14): 1137-1143.
- Lv M, Xu P, Wu Y, Huang L, Li W, et al. (2016) LncRNAs as new biomarkers to differentiate triple negative breast cancer from non-triple negative breast cancer[J]. Oncotarget 7: 13047-13059.
- Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY (2013) Analysis of long noncoding RNA expression profifiles in gastric cancer[J]. World J Gastroenterol 19: 3658-3664.
- Han L, Li Z, Jiang Y, Zheng Jiang, Ling Tang (2019) SNHG29 regulates miR-223-3p/CTNND1 axis to promote glioblastoma progression via Wnt/β-catenin signaling pathway[J]. Cancer Cell Int 19: 345.
- Wang, Tian, Zhou, Guangming Wang, Wenlei Yu, et al. (2018) A threelncRNA signature for prognosis prediction of acute myeloid leukemia in patients[J]. Molecular Medicine Reports 18(2):1473-1484.

 Research Article
Research Article