Synopsis
Human Papillomavirus (HPV) DNA test improved the effectiveness of Cervical
Cancer Screening Programs, but it frequently leads to unnecessary follow-up and
treatments in potentially regressing lesions. The presence of HPV oncoproteins, named
E6 and E7, is a key event resulting from persistent infection, besides other factors are
certainly involved. Proteomics represents a promising tool to precociously identify the
possible changes occurring during cervical carcinogenesis and to better understand
the natural history of HPV-related cancer. The suitability of PreservCyt medium for gelbased
proteomic analysis has not been tested yet. This study aimed to build a healthy
cervix reference map by two-dimensional Matrix-Assisted Laser Desorption/Ionization
time-of-flight mass spectrometry (2DE MALDI-TOF MS) starting from Thin Prep cervical
specimens. The first step was to compare residual and whole specimens in terms of
protein recovery, the second being the assessment of the influence of storage duration
on proteomic analysis.
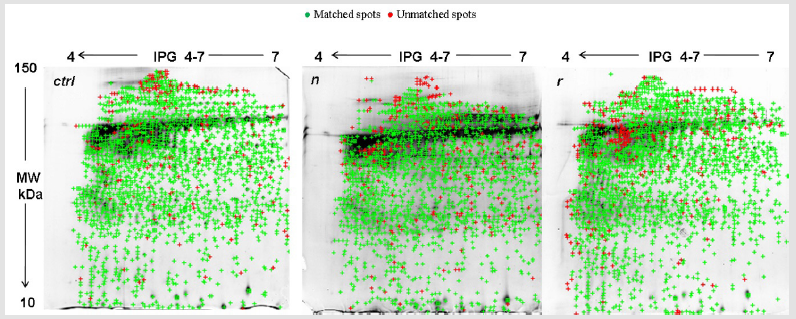
Image analysis of 2DE gel identified a similarity of 87% between residual and whole
samples. Protein recovery drastically decreased when storage duration increased. Many
of the proteins identified in study samples are constitutively present in healthy cervix.
Thin Prep cervical samples are suitable for proteomic analysis. The “one sample for all”
approach confirms to be possible.
Keywords: Cervical Cancer; Biobanks; 2DE gel-based proteomics; 2DE map; Preserv- Cyt solution; Thin Prep
Introduction
Human Papillomavirus (HPV) infection represents a necessary
but far from sufficient event during cervical carcinogenesis since
multiple steps are necessary to drive cells towards invasive cancer.
The introduction of HPV-DNA test in the diagnostic algorithm of
patients with cervical lesions improved the effectiveness of cervical
cancer screening. DNA testing demonstrated high sensitivity in
detecting Cervical Intraepithelial Neoplasia (CIN), thus reducing
cervical cancer incidence [1]. However, the main problem with HPVDNA
test remains the high prevalence of HPV infection in comparison
with the low percentage of women developing cancer [2]. The low
specificity of HPV-DNA test does not distinguish between transient
infections, usually clearing in two years, and persistent infections
that harbour the highest risk of progression toward cancer [2]. The
better doing to prevent Cervical Carcinoma (CC) is to recognize
intraepithelial lesions needing medical interventions without
concerning HPV infections which are transitory. It is therefore
important to focus on supplementary tests, which can limit overtreatments
by giving additional information about the outcome of
the lesions [3].
Viral persistence is often caused by loss of immune regulation
that drives cell towards disruption of cell-cycle control and
accumulation of neoplastic events [4]. Unregulated HPV E6/E7 genes represents a key event, but other factors are involved
during neoplastic switch. Thus, the global understanding of the
molecular mechanisms Underlying the progression of cervical
intraepithelial lesions and their prognosis should be improved.
“Omics” technologies rely on the holistic view of an organism as a
puzzle of molecules. If considered as a whole, the human system
can be systematically better understood [5]. Proteome is defined
as the set of all protein species in a cell, tissue or organism under
physiological conditions. Thus, changes at the protein level would
reflect pathological conditions and their severity. In this context,
proteomic analysis, showing differential protein patterns in the
various steps of cervical carcinogenesis, would certainly clarify
the intricate interplay existing between HPV infection and human
cervix. Ensuring a proteome identity for each individual patient
with HPV infection would mean to provide a tailored strategy for
diagnosis, clinical management and therapy [6,7].
Based on those premises, high quality samples are essential for successful results. Biobanks are founded on the systematic storage of human specimens and associated data. Nowadays they are considered as useful platforms for personalized medicine, offering the best way to translate data from research to clinical practice [7]. In particular, Cervical Cytology Biobank (CCB) could be precious to maximize proteomic studies about cervical carcinogenesis [8]. Molecular analyses on cytological samples are currently feasible because of the introduction of Liquid-based Cytology (LBC) [9]. The main advantage of LBC is that additional tests can be run on residual cells of a single sample [9,10]. A residual Thin Prep specimen (Hologic Inc., Marlborough, MA) is a PreservCyt solution (Hologic Inc., Marlborough, MA), containing remaining cells after the conclusion of the routine screening round. Thin Prep represents the most widely used Liquid Based (LB) Pap test among the two FDA-approved ones [10]. PreservCyt solution is a methanol-based fixative medium [10].
Differential protein pathways during the consecutive steps of cervical carcinogenesis have been investigated on tissues and plasma samples [11,12]. Data demonstrating the possible use of ethanol-fixed tissues for proteomic analyses also exist [13,14]. Papachristou et al. reported a non-targeted analysis of HPV protein in PreservCyt specimens by shotgun analysis [15]. To our knowledge, no one explored the suitability of PreservCyt medium to build a gel-based 2DE reference map for healthy cervix. The main goal of our research was to find the possible changes occurring in cervical proteome during HPV-related carcinogenesis, in order to integrate proteome profiling with genome data. Therefore, in this study we checked the feasibility of residual Thin Prep cervical specimens as potential source for gel-based proteomic analysis, and tried to set up a reference 2DE map of human healthy cervix to identify putative predictive biomarker for CC.
Material and Methods
Sample Collection
The present retrospective incidence-based study was performed by collecting data from the electronic regional cervical cancer registry linked to the regional tissues and cells Biobank, named Adriatic Biobank (AB). AB is placed in the Surgical Pathology Unit of the University Hospital of Chieti. At room temperature (RT), AB stores about 50,000 residual cervical specimens collected from women participating in Cervical Cancer Screening Program. During gynaecological examination, cervical specimens were taken from ecto-endocervix by Cervex Brush (Rovers Medical Devices, Oss, Netherlands) and dispersed in 20 mL of PreservCyt solution. To assess the feasibility of PreservCyt specimens for proteomic analyses, we compared time zero samples, consisting of cervical cells stored in PreservCyt medium for approximately two hours and whose aliquots were taken before undergoing routinely screening round (henceforth named naive, n) [9] with fresh frozen samples (untreated sample, ctrl), consisting of cells removed from patients during hysterectomy, immediately thus transferred to dry ice, within 30 min to a -80 °C freezer.
To investigate the possible influence of specimen handling on the protein recovery, we compared n specimens with residual ones (r), the latest corresponding to samples completing routine screening round [9]. In our Institution, a routine screening round for HPV negative cases is usually completed in two days. Finally, the influence of storage duration on protein recovery was tested on six time-groups of specimens as follows: two hours (n), two days (r), two months (2m), six months (6m), one year (1y) and four years (4y). For each group we selected three samples as biological replicates. The exclusion criteria for samples selection were history of any interventions for CIN in the previous five years, chemotherapy/radiotherapy, hysterectomy, pregnancy, immunodeficiency diseases. To avoid bias only specimens related to patients with standardized physiological conditions were used.
Sample Processing, 2DE Preparation And Gel Analysis
An aliquot of 3.5x104 cells for each sample, corresponding to about 5 ml of cellular material [9,10], were centrifuged at 2000 rpm for 5 min at 4°C. The pellet was washed with PBS 1x, then resuspended in 8M urea, 40mM Trizma Base and 4% CHAPS (Reagent 1 Merck, KGaA, Darmstadt, Germany) and supplemented with 1% Tributylphosphine and 1% Protease Inibitor Mix. Once sonicated, each sample was incubated with endonuclease (500 U) for 10 minutes at RT and centrifuged at 14800 rpm for 15 min at 4°C. Proteins were precipitated from supernatant with 3 volumes of ice cold acetone overnight at -20°C and next centrifuged at 14800 rpm for 15 min. Proteins concentration was measured using the Pierce Coomassie Plus (Bradford) Protein Assay kit (Thermo Fisher Scientific, Waltham, MA US), with BSA, as standard. 150 μg and 500 μg of each sample for analytical and preparative gel, respectively, were mixed with rehydration solution (DeStreak Rehydration Solution, GE Healthcare, Uppsala, Sweden). To separate the mixture of proteins into individual protein spots, according to their pI and MW, rehydrated protein extracts were separated by 12% acrylamide 2DE gel. Isoelectric focusing (IEF, Ettan IPGphor III System GE Healthcare) was applied using 18 cm IPG strips (GE Healthcare, Uppsala, Sweden) and covering a pH range of 4-7. Analytical gels were stained with ammoniacal silver nitrate, while gels used for protein identification by MALDI-TOF MS were silver-stained without glutaraldehyde, in accordance to the mass compatible method described by Shina et al. [16].
Three different gels were run for each sample and conditions (technical replicates). On gels, proteins appeared as spots which subsequently undergoing image analysis by Image Master 2D Platinum 6.0 software (GE Healthcare, Uppsala, Sweden). The software detected the different spots basing on optical density of silver-stained protein. A positional gel calibration was carried out using the 2D calibration method included in the analysis package that calculated the position of protein spots in terms of pI and MW values. Before digital analysis of the different spots, some landmarks (Lms) were manually identified. Next, 2DE gels were analysed, digitized and data were exported as TIFF files.
MS proteins Identification
All the common proteins obtained from the matching spots and whose intensity levels were significantly high (p value < 0.05), were picked manually and analysed through Peptide Mass Fingerprinting (PMF) approach with MALDI-TOF MS. A spot was considered significant only if it was detected in three out of all technical replicates for each sample. Following protein spot digestion in a 50 mM NH4HCO3 buffer containing trypsin, and incubation at 37 °C overnight, the peptide extract was applied to a C18 ZipTip pipette tips (Millipore, CA, USA), rinsed with a 0.1% TFA and eluted directly on the MALDI target with 0.5 μl of a saturated α-cyano- 4-hydroxycinnamic acid (1:1 = ACN: 0.1% TFA) solution. Tryptic digests were analyzed by Autoflex™ Speed mass spectrometer (Bruker Daltonics, Bremen, Germany) equipped with a Nd:YAG laser (355 nm; 1000 Hz) operated by FlexControl v3.3, equipped with a 355-nm nitrogen laser. All spectra were obtained with the delayed extraction technology in positive reflectron mode and averaged from 100 laser shots to improve the S/N ratio. Samples were analysed through PMF and examined using LIFT MS/MS from the same target. The most abundant ions for each sample were chosen for MS/MS analysis. Analyses were performed in positive LIFT reflection mode.
Precursor ion selector range was 0.65% of parent ion mass. After MS acquisition, each spectrum was submitted to PMF to to be searched in NCB protein database using a Mascot search engine. Mass tolerance of 100 ppm for PMF and 0.6-0.8 Da for MS/ MS was used. Following automated assessment of search results, the samples were automatically submitted to LIFT TOF/TOF acquisition for validation of data from PMF. A maximum of four precursor ions per sample were chosen for MS/MS analysis. Mascot database search analyses using combined PMF and MS/MS datasets were performed via BioTools 3.2 (Bruker Daltonics) connected to the Mascot search engine used for SwissProt database search of datasets. A score of about 70 for PMF and 30-40 for MS/MS search was considered as the identity threshold.
Bioinformatics Analysis
Protein ontology classification was performed using Gene Ontology system [17]. Proteins were grouped according to their associated biological processes, cellular localization and molecular pathway, using a multi-staged classification methodology based upon 13.1 “PANTHER” (http://www.pantherdb.org/, SRI International, Menlo Park, CA) software [18]. STRING software was utilized for qualitative and quantitative comparison of protein spots [19]. To determine the reproducibility of this proteomic platform, three technical replicates were analysed. Final data were collected in February 2020.
Statistical Analyses
Pearson’s index investigated the correlation between storage duration and protein recovery. ANOVA test evaluated differences between independent variables. Statistical tests were performed by SPSS software (15.0 SPSS for windows, Inc., Chicago, IL) regarding probability values p less than 0.05 as statistically significant.
Ethical Aspects
The study was approved by G. d’Annunzio University of Chieti-
Pescara and Local-Health-Authority. The prevailing norms of Good
Clinical Practice were respected in accordance with 2013 Helsinki
Declaration.
Before gynaecologic examination, each woman signed a written
informed consent accepting the use of residual specimen for
scientific research. As for confidentiality standards, an identification
code was assigned to each specimen collected in Adriatik Biobank.
Results
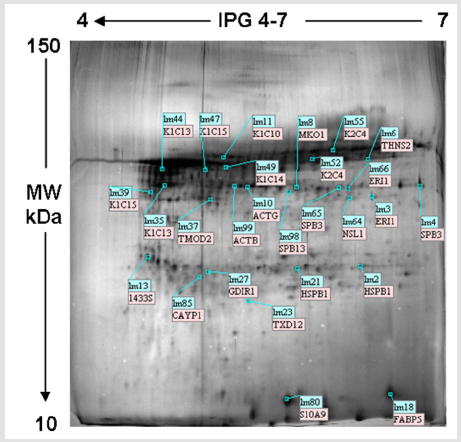
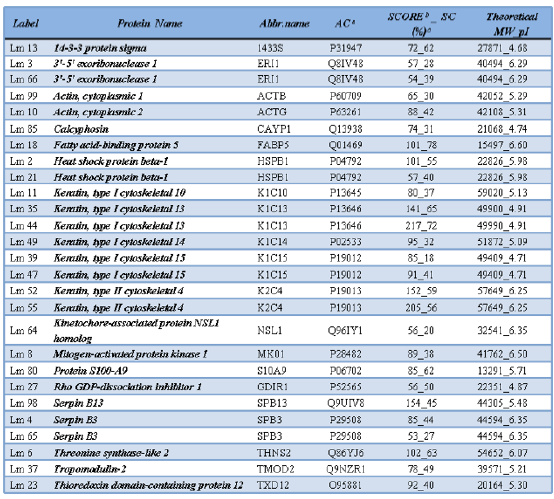
The selected specimens were taken from women ranging from 25 to 64 years (median 40.3±15), reporting no cytological abnormalities and resulting as HPV-DNA negative. Protein profile visualised a mean of 1941±85 spots for ctrl, 2072±95 for n and 2086±101 for r samples. Differences were not significant (p=0.9). 2DE gel analysis revealed a median value of 1837±58 for matched spots, and 271±11 for unmatched ones, with an 87% similarity (Figure 1). Among technical and biological replicates, the reproducibility was about 85%, thus reflecting the small variation among samples and runnings. Only matched spots, called Lms, underwent further analysis. Low scoring peptides were not considered. Twenty-seven of the most abundant protein were excised from spots and evaluated by MALDI-TOF MS. Among these, six proteins were classified as isoforms, differing in isoelectric point and/or molecular weight. Molecular mass of the excised proteins ranged from 10 to 150 kDa. The isoelectric point was in the 4-7 pH range. Gel localization, name and Lms of the proteins assigned to each spot are illustrated in Figure 2. Table 1 lists the twenty-seven identified proteins (including isoforms) and the related information obtained by databases. P value was based on Swiss Prot database using the MASCOT searching program as MALDI-TOF MS data.
Figure 1: 2DE gel comparison of total protein extracts between fresh frozen (ctrl), naive (n) and residual (r) samples. 2DE, two-dimensional gel electrophoresis.
Figure 2: Typical 2DE profile of residual sample proteins showing the 27 identified proteins indicated by Lms. Lms, landmarks.
Table 1: List of the 27 detected proteins identified by MS.
aAC, access code number from Swiss Prot database.
bScore: -10*Log(P), where P is the probability that the observed match is a random event.
cSC%, sequence coverage: ratio between the portion sequence covered by matched peptide and the full length of the protein sequence.
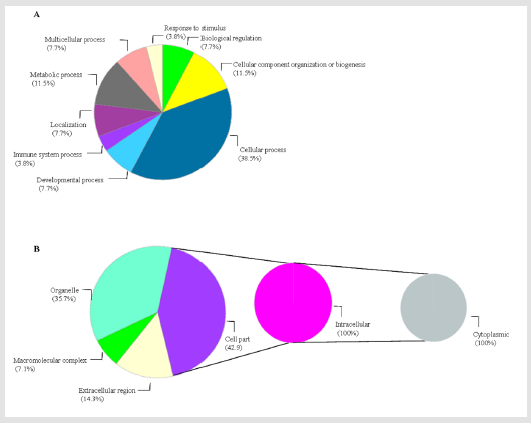
Proteins were classified according to biological processes,
cellular localization and protein classes. Regarding biological
process, proteins implicated in cellular processes (38.5%), cellular
component organization and biogenesis (11.5%), metabolic
process (11.5%), developmental process (7.7%) and immunity/
defense (3.8%) represent the largest categories (Figure 3A). Among
the 42.9 % of cell part proteins, 100% were intracellular types
with cytoplasm localization (Figure 3B). Cytoskeletal proteins and
enzyme modulators were the most represented molecules (23.1%),
followed by signalling molecules (15.4%). Bioinformatics analysis revealed that the vast majority of cytoskeletal proteins were actinassociated
proteins and citokeratins. Among enzyme modulators,
Serpin B and Rho GDP-dissociation inhibitor were detected,
including S100A9 signalling protein. Both “Inflammation mediated
by chemokines and cytokines” (6.4%) and “integrin signalling”
(6.4%) were the predominant patterns. Both the above-mentioned
pathways included proteins belonging to actin family.
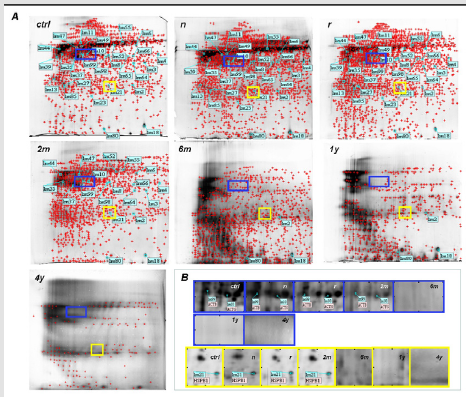
In 2DE maps obtained from ctrl, n, r, 2m, 6m, 1y and 4 y samples,
a median number of 1941±85, 2072±95, 2086±101, 1285±32,
975±52, 518±34 and 215±32 spots were identified, respectively
(Figure 4A). A strong correlation between storage duration and
proteins recovery has been found (R=0.89). Differences were
statistically significant (ANOVA test, p<0.05). At the opposite
side, difference among biological and technical replicates was
not significant. Since loading was balanced in terms of number of
cells for sample, the colour yield of each spot changed in relation
to storage duration. Gel zooming of selected spots chosen as the
examples, demonstrated the progressive loss of proteins integrity
along time, until spots disappearance in gel from samples 6m, 1y
and 4y (Figure 4B). 2DE analysis was performed considering ctrl
samples as the benchmark. The mean percentage of matching spots
decreased with the increasing of storage duration, as follows: 67 %
for n, 60% for r, 50% for 2m, 38% for 6m, 18.9% for 1y and 1.3 for
4y. Differences reached statistical significance (p<0.05).
Figure 4: A. Comparison of 2DE gels of the total proteins extracts from fresh frozen specimens (ctrl) and PreservCyt samples having different storage duration (n, r, 2m, 6m, 1y and 4y). B. Gel zooming of Lms spots 99 and 10 (blue rectangle) and 21 (yellow square) to demonstrate the progressive decreasing of proteome integrity along with the increasing of PreservCyt storage duration.
Discussion
This study represents the first gel-based proteomic analysis carried out on residual Thin Prep cervical samples. One of the greatest challenges in the modern medicine is to deeply understand the cascade of molecular events leading to tumour development and tumour progression. Nowadays, the accurate prediction of tumour outcome represents one of the main issues for clinical oncology. In this context, proteome analysis achieved impressing results. Proteome characterization, evaluating protein species, molecular functions and biological processes, provides a complete view about tumour biology [20], thus making possible to translate evidencebased medicine from the bench to the bedside. Over the recent years, tremendous technological innovation has been achieved in protein field, such as the SDS-PAGE approach via gel-based LC/MS [21]. Each cell contains most proteins. Each protein is often present like different protein species, at least some of them separated by 2DE. Each protein species represents a different chemical molecule with a potential different function. Using 2DE MS approach it is possible to obtain relevant results, in terms of protein function. [22]. This potentiality clearly exceeds the power of the bottom-up LC-MS approach alone [22].
An adequate sample preparation is crucial to standardize and reproduce 2DE protocols. An attempt in identifying potential protein biomarkers for cervical cancer was carried out on plasma samples. The results showed low specificity, due to plasma contact with all the organs of the body [23]. Gu et al., also aim to identify candidate protein biomarkers for high-grade dysplastic cervical cells using mass spectrometry, but they used a different approach. They isolated both normal and high-grade cervical cells starting from Thin Prep slides and using a Laser Capture Microdissection. To reach approximately 12,000 cells, they pooled them from slides belonging to different cases [24]. Aiming to develop a protocol for obtaining high-quality protein from a simple and cheap diagnostic tool as the liquid-based Pap test is, we compared fresh frozen cytology samples with PreservCyt specimens, and demonstrated the suitability, feasibility and efficiency of Thin Prep Pap test for the identification of proteins by 2DE and MALDI-TOF MS combination.
The high matching rate detected between r and n specimens would suggest that the handling of LB samples for routinely diagnostic purposes does not affect proteins recovery. On the other hand, and in contrast with our previous data demonstrating the stability of nucleic acids in residual LB specimens for a long time [25], the decreasing number of spots detected in older samples suggests that long storage duration negatively influences proteome integrity. In this context, the change in colour yield of the single spot in relation to storage duration would support our hypothesis. Loss of protein integrity would probably be a consequence of the timedepending modification of protein structure by chemical crosslinking [26] or is probably due to the progressive consumption of methanol-based fixative medium overtime. It’s our opinion that a reasonable and precautionary cut-off between acceptable and unacceptable storage duration should be set at two days. A promising solution to by-pass this issue would rely on the use of aliquots of PreservCyt specimens taken within two days from collection and stored at -80°C [25, 27, 28]. This approach would be certainly easy to achieve in clinical settings too.
Among the wide range of molecular pathways detected in our samples, “inflammation mediated by chemokines (KFs) and cytokine signaling” were the most represented. Cervical cells physiologically secrete natural microbicides, mucus, and a mixture of peptides, such as chemokines and cytokines, which are able to regulate the traffic and activity of immune cells. These mediators, which are unanimously considered as the physiological components of cervical milieu, would contribute to the homeostasis of the female reproductive tract, uterine health and reproduction [26]. KFs and actins (actin beta and actin gamma) resulted as the main components of 2DE map obtained by healthy cervix. Cytoskeleton is a network of fibers by which cells are supported and maintain their shape. It is also involved in cell division, motility, protein sorting and signal transduction [29]. Human KFs comprise a large family of proteins, numbered 1-20, distributed in a tissue-specific manner and differentiation-related way. They contribute to cell type-specific functions, such as adhesion, migration, and metabolism [30].
KFs are unanimously considered as epithelia differentiation markers. Although many types of KFs are commonly considered as contaminants in MS, those identified in our study have to be regarded as typical for ectocervical squamous epithelium [31]. In particular, keratin 4 type II is present in the basal cell layer, keratins 13 and 14 are characteristic of reserve cells, and keratin 10 is distinctive for suprabasal cell layer. In CIN3, reserve cells lose keratins 13 and 14 while acquiring keratins 8, 17 and 18 [32]. Neoplastic cells commonly change their morphology with reduction of cell adhesion due to cytoskeletal disruption. Cell migration is strictly regulated by the re-organization of the actin fibers. In nonmuscle cells, actin cytoskeleton is represented by α and β isoforms [33]. Liao et al., demonstrated that HPV16/18 E5 would contribute to cancer progression through the regulation of actin cytoskeleton network [34].
Serpin B3 (SPB3) is a serine protease inhibitors belonging to the serpin superfamily. It is located in the spinous and granular layers of the normal squamous epithelium of several organs and is physiologically involved in the regulation of cells differentiation [35]. SPB3 level is high in cervix [36]. Serpins B13 is a cathepsin G inhibitor. Cathepsin G was detected within cervicovaginal fluid where it is chemoattractant for monocytes/neutrophils and stimulates T-cells [35]. Serpins B13 is constitutively present in healthy cervix and would protect lower genital tract from inflammation [36].
14-3-3 sigma protein (1433 S) was recognized as one of the major G2/M checkpoint control gene. It is inactivated in some types of cancer mainly through epigenetic hypermethylation [37]. Holm et al. reported the presence of 1433S protein in the cytoplasm of healthy cervical cells [38]. In their study carried on cervical histological specimens from women diagnosed with HPV infection, Syrjänen et al. demonstrated an increased presence of 14-3-3 sigma that paralleled with the grade of CINs [39].
S100A9 belongs to the S100 family of calcium-binding proteins. S100A9 regulates cell growth, differentiation, migration, adhesion and signal transduction. In Zhu et al., S100A9 improved from healthy cervix to CIN [40].
Fatty acid binding proteins (FABPs) are a family of small, cytoplasmic proteins playing a role in fatty acid uptake, transport, and metabolism. They are present in cervical epithelial cells. FABP5 is up regulated in cervical cancer, where it would play a crucial role in cancer progression, invasion, and metastasis. Particularly, it would acts through the reprogramming of fatty acid metabolism, the regulation of angiogenic responses, and the promotion of lymphangiogenesis. FABP5 would also participate in epithelial-to-mesenchymal transition, that is considered the initial stage of metastatic progression and is characterized by specific morphogenetic changes, loss-of cell-cell adhesion, and increased cell motility [41]. Heat shock proteins (HSP) are molecular chaperones which are constitutively present in cells. They are able to protect cells from various stresses, such as high temperature, anoxia or chemical agents. Heat shock protein beta 1 (HSPB1), also named as heat shock protein 27 (Hsp27), is induced by environmental stress and developmental changes. It moves from the cytoplasm to the nucleus of the cell upon stress induction; moreover, it is involved in actin organization. HSPB1/Hsp27 is commonly present in cervical epithelial cells and ensures cell death resistance in HeLa cells [42].
Kinetochore-associated protein NSL1 homolog belongs to
a complex of proteins which are able to bridge centromeric
heterochromatin with the outer kinetochore structure. NSL1
homolog is a constitutive protein of the healthy cervix; it is up
regulated in cervical squamous epithelium.
The mitogen-activated protein kinase (MAPK1/MK01) is a
serine-threonine kinase that is highly conserved during biological
evolution. MAPK1/MK0 is able to independently transduce the
extracellular signals into cells, in order to regulate the related genes. It is also involved in physiological processes such as
mitosis, proliferation, inflammation, invasion and metastasis
[43]. The presence of MAPK1 in cancer tissue was found to be
higher with respect to the adjacent healthy tissue [44]. A model of
cervical tumorigenesis induced by the E6 oncogene validated the
involvement of the mitogen-activated protein kinase signalling via
Ras oncogene [45].
Rho GDP dissociation inhibitor protein 1 (GDIR1) is a protein
signature of the healthy cervix. On the other side, it is downregulated
in cervical cancer in which it mediates migration, attachment and
organization of cells into tissues [46]. In mice, threonine synthaselike
protein (THNS2) catalyses the degradation of O-phosphohomoserine
to alpha-ketobutyrate, phosphate and ammonia. An
alternatively spliced form of this protein has been shown to act as a
cytokine inducing inflammation [47].
Tropomodulin 2 (TMOD2) is a member of the tropomodulin
family, considered as actin-regulatory proteins. TMOD2 caps the
pointed end of actin filaments preventing both elongation and
depolymerisation [48].
Finally, the thioredoxin system maintains the reduced state of
intracellular environment. It is also involved in the modulation of
cell growth/cell death mechanisms and in the oxidized proteins
repair. Thioredoxins are present in human cervix and are up
regulated in a variety of human cancers, including cervical one.
Being the predominance of thioredoxins linked to aggressive
tumour growth and poor prognosis, these molecules have recently
been considered as important targets for cancer therapy [49].
The detailed examination of some of the proteins detected in our
study, most of which are constitutively present in healthy cervix and
certainly modulated during cervical carcinogenesis, would confirm
that residual PreservCyt-based samples contain a sufficient amount
of proteins for 2DE/ MS analysis. Thus, Thin Prep Pap test have to
be considered a valuable and precious source to draw a tailored
approach to improve the effectiveness and appropriateness of
cervical cancer prevention strategies. The availability of a medium
that is routinely used in clinical settings would undoubtedly be
realistic, with practical benefits for healthcare services. On the
other hand, storage duration should be carefully regarded. The “one
sample for all” approach confirms to be possible [25].
References
- Ronco G, Dillner J,Elfström KM, Sara T, Peter JFS, et al. (2014) Efficacy of HPV-based screening for prevention of invasive cervical cancer. Follow-up of four European randomized controlled trials. Lancet 383:524-532.
- Maggino T, Sciarrone R, Murer B, Maraia Rossi, Chaira F, et al. (2016)Screening women for cervical cancer carcinoma with a HPV mRNA test: First results from the Venice pilot program.BrJCancer 23: 525-532.
- Zappacosta R, Rosini S(2008) Cervical cancer screening: from molecular basis to diagnostic practice, going through new technologies. Technol Cancer ResTreat7(3): 161-174.
- Hoppe Seyler K, Bossler F,Braun JA, Herrmann AL, Seyler FH (2018) The HPV E6/E7 Oncogenes: Key Factors for Viral Carcinogenesis and Therapeutic Targets. Trends Microbiol26(2):158-168.
- Boja ES, Kinsinger CR, Rodriguez R, Srinivas P, behalf of Omics Integration Workshop Participants. (2014) Integration of omics sciences to advance biology and medicine.ClinProteomics 11(1): 45-56.
- Boda D, Docea AO, Calina D, C Caruntu, Llie Ghita MA, et al. (2018) Human papilloma virus: Apprehending the link with carcinogenesis and unveiling new research avenues.Int JOncology 52:637-655.
- Kinkorovaà J (2015) Biobanks in the era of personalized medicine: objectives, challenges, and innovation: Overview. EPMA Journal 7(1):4-9.
- Castle PE, Solomon D, Hildesheim A, Ronaldo H, C Bratti, et al. (2003) Stability of archived liquid-based cervical cytologic specimens. Cancer 99:89-96.
- McGoogan E (2006) Liquid-based cytology. In Monsonego editor, Emerging Issues on HPV Infections: From Science to Practice. 1stednBasel: Kargerp147-156.
- Hologic (2010) Thin Prep PreservCyt solution material safety data sheet. Hologic Inc, Bedford.
- SerafínHiguera I, GaribayCerdenares OL, IlladesAguiar B, Eugenia Flores, Marco Antonio, et al. (2016) Differential proteins among normal cervix cells and cervical cancer cells with HPV-16 infection, through mass spectrometry-based Proteomics (2D-DIGE) in women from Southern Mé Proteome Science 14(10):2-9.
- Xia G, Guzalnur A, Reyimu H, Feng Zhao, N Kadeer, et al. (2002) The association of a distinct plasma proteomic profile with the cervical high-grade squamous intraepithelial lesion of Uyghur women: a 2D liquid-phase chromatography/mass spectrometry study. Biomarkers17(4):352-361.
- Ahram M, Flaig MJ, Gillespie JW, Paul HD, W Martson, et al. (2003) Evaluation of ethanol-fixed, paraffin-embedded tissues for proteomic applications. Proteomics 3(4):413-421.
- Chaurand P, Latham JC, Lane KB, James AM, Vasily PA, et al. (2008) Imaging Mass Spectrometry of Intact Proteins from Alcohol-Preserved Tissue Specimens: Bypassing Formalin Fixation. JProteome Research 7(8):3543-3555.
- Papachristou EK, Roumeliotis TI, Chrysagi A Trigoni C, E Charvalos, et al. (2013) The shotgun proteomic study of the human ThinPrep cervical smear using It RAQ mass-tagging and 2D LC-FT-Orbitrap-MS: the detection of the human papillomavirus at the protein level. Journal of Proteome Research 12:2078-2089.
- Shiha P, Poland J, Schnolzer M, Rabilloud T(2001) A new silver staining apparatus and procedure for matrix-assisted laser desorption/ionization-time of flight analysis of proteins after two-dimensional electrophoresis. Proteomics 1(7):835-840.
- Harris MA, Clark J, Ireland A, Jane L, Rebecca F, et al. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Research 32:258-261.
- Mi H, Muruganujan A, Thomas PD (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Research 41:377-386.
- Szklarczyk D, Morris JD, Cook H, Michael K (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research 4(45):362-368.
- Schlüter H, Apweiler R, Holzhütter RHG, Jungblut PR (2009) Inding one's way in proteomics: a protein species nomenclature.Chem Cent J 3(1):11.
- Giannopoulou GE, Garbis SD, Vlahou A, Kossida S, Lepouras G, et al. (2009) Proteomic feature maps: a new visualization approach in proteomics analysis. Journal of Biomedical Informatics 42: 644-653.
- Zhan X, Yang H, Peng F, Jianglin L, Yun M, et al. (2018) How many proteins can be identified in a 2DE gel spot within an analysis of a complex human cancer tissue proteome? Electrophoresis 39:965-980.
- Guo X, Hao Y, Kamilijiang M, Hasimu A, Yuan J, et al. (2015) Potential predictive plasma biomarkers for cervical cancer by 2D-DIGE proteomics and Ingenuity Pathway Analysis. Tumor biology 36(3):1711-1720.
- Gu Y, Wu SL, Meyer JL, Williams SH, Lawrence JB, et al. (2007) Proteomic analysis of high-grade dysplastic cervical cells obtained from ThinPrep slides using laser capture microdissection and mass spectrometry. Journal of Proteome Research 6:4256-4268.
- Zappacosta R, Sablone F, Pansa L, Buca, Rosini S, et al. (2017) Analytic and Diagnostic Performances of Human Papillomavirus E6/E7 mRNA Test on up-to 11-Year-Old Liquid-Based Cervical Samples. A Biobank-Based Longitudinal Study.IntJMolSci18:1480-1495.
- Fahey V, Schaefer TM, Channon JY, Wira CR (2005) Secretion of cytokines and chemokines by polarized human epithelial cells from the female reproductive tract. Human Reproduction 20(6):1439-1446.
- Hu S, Xie Y, Ramachandran P(2005) Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics 5:1714-1728.
- Chevalier F, Hirtz C, Chay S, F Cuisiner, Sormmerer N, et al. (2007) Proteomic Studies of Saliva: A Proposal for a Standardized Handling of Clinical Samples. Clin Proteom 3:13-21.
- Lee SH, Dominguez R (2010) Regulation of actin cytoskeleton dynamics in cells. MolCells 29:311-325.
- Depianto D, Kerns ML, Dlugosz AA, Coulombe PA (2010) Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat Genet 42:910-914.
- Zegels G, Van Raemdonck GA, Coen EP, Tjalma WA, Van Ostade XW (2009) Comprehensive proteomic analysis of human cervical-vaginal fluid using colposcopy samples. Proteome Sci 7:17-23.
- Efremov YM, Dokrunova AA, Efremenko AV, Kirpichnikov MP, Shaitan KV, et al. (2015) Distinct impact of targeted actin cytoskeleton reorganization on mechanical properties of normal and malignant cells. BiochimBiophys Acta 1853:3117-3125.
- Simiczyjew A, Mazur AJ, Dratkiewicz E, Nowak D (2017) Involvement of β- and γ-actin isoforms in actin cytoskeleton organization and migration abilities of bleb-forming human colon cancer cells. Plos One 23:1-22.
- Liao S, Deng D, Zhang W, Xiaoji Hu, Wei Wang, et al. (2013) Human papillomavirus 16/18 E5 promotes cervical cancer cell proliferation, migration and invasion in vitro and accelerates tumor growth in vivo. Oncol Rep29:95-102.
- Burgener A, Boutilier J, Wachihi C, J Kimani, M Carpenter, et al.(2008) Identification of Differentially Expressed Proteins in the Cervical Mucosa of HIV-1-Resistant Sex Workers. JProtRes 7:4446-4454.
- Curato C, Pontisso P (2015) SERPINB3 (serpin peptidase inhibitor, clade B (ovalbumin), member 3. Atlas Genet. CytogenetOncolHaematol 19(3): 202-209.
- Sano T, Shimooka H, Weixa P, Segawa A, Jian Z, et al. (2004) Immunohistochemical expression of 14-3-3 sigma protein in various histological subtypes of uterine cervical cancers. PatholInt 54(10):743-750.
- Holm R, Ali T, Svendsrud DH, Nesland JM, Kristensen GB, et al. (2009) Expression of 14-3-3sigma in cervical squamous cell carcinomas. OncolRep 22:11-15.
- Syrjänen S, Naud P, Sarian L, Sophie D, Ahemar LF, et al.(2010) Up-regulation of 14-3-3sigma (Stratifin) is associated with high-grade CIN and high-risk human papillomavirus (HPV) at baseline but does not predict outcomes of HR-HPV infections or incident CIN in the LAMS study. AmJClinPathol 133:232-240.
- Zhu XJ, Zheng Y, Zhou SW, Jiang W, Jiang L, et al. (2010) Expression and significance of S100A9 in progression of squamous cervical cancer. JPractMed26(8):1350-1352.
- Wang W, Chu HJ, Liang YC, Huang JM, Chang CL, et al. (2016) FABP5 correlates with poor prognosis and promotes tumor cell growth and metastasis in cervical cancer. TumorBiol 7(11):14873-14883.
- Real NE, Castro GN, Darío CuelloCarrión F, Claudia P, Hanna R, et al. (2017) Molecular markers of DNA damage and repair in cervical cancer patients treated with cisplatin neoadjuvant chemotherapy: an exploratory study Cell Stress. Chaperones 22(6): 811-822.
- Huang C, Liu LY, Li ZF, Wang P, Lei N, et al. (2008) Effects of small interfering RNAs targeting MAPK1 on gene expression profile in HeLa cells as revealed by microarray analysis. CellBiolInt 32(9):1081-1090.
- Li XW, Tuergan M, Abuliz G (2015) Expression of MAPK1 in cervical cancer and effect ofMAPK1 gene silencing on epithelial-mesenchymal transition, invasion and metastasis. Asian PacifJTropMed 8(11):937-943.
- Jang M, JeeEun Rhee JE, Jang DH, Kim SS (2011)Gene Expression Profiles are Altered in Human Papillomavirus-16 E6 D25E-Expressing Cell Lines. VirolJ 8:45345-45349.
- Bae SM, Min HJ, Ding GH, Kwak SY, Chao YL, et al. (2006) Protein Expression Profile using Two-Dimensional Gel Analysis in Squamous Cervical Cancer Patients. Cancer ResTreat 38(2):99-107.
- (2016) Harmonizome, Threonine synthase-like 2 (S. cerevisiae).THNSL2 Gene:55258.
- Omotade OF, Rui Y, Lei W, Kuai Y, Criss H, et al. (2018) Tropomodulin Isoform-Specific Regulation of Dendrite Development and Synapse Formation. J Neurosci38(48):10271-10285.
- Tibodeau JD, Benson LM, Isham CR, Owen WG, Bible KC (2009) The anticancer agent chaetocin is a competitive substrate and inhibitor of thioredoxin reductase. AntioxidRedox Signal 11(5):1097-1106.

 Research Article
Research Article