Abstract
There is consensus that SARS-CoV-2 originated in horseshoe bats as it shares a high sequence similarity to bat CoVs, ~96% with the bat-CoV-RaTG13 and ~90% with bat-SL-CoVZC45 and bat-SL-CoVZXC21. But, the bat CoVs are not its direct ancestors, instead an intermediate host is involved, which subsequently transferred the virus to humans. The Malayan pangolin harbouring CoVs with a high similarity to SARS-CoV-2, is the most potential intermediate host. There is another consensus, that SARS-CoV-2 has higher infectivity and transmissibility as compared to SARS-CoV, related to a unique peptide (PRRA), four amino acid residue insertion on the S protein between its S1 and S2 subunits and involved in the proteolytic cleavage of the spike protein by cellular furin-like proteases widely expressed in lungs and respiratory tract and various other organs. This furin cleavage site is distinct from SARS-CoV and other CoVs including pangolin CoV which contain only a trypsin or TMPRSS2 cleavage site. This trait, the novel furin cleavage site on the S protein in SRS-CoV-2, has helped the its spill-over to humans and potentiated its transmissibility as well as pathogenicity.
The Perspective and Implications
The sequences of SARS‐CoV‐2 from various patients from different countries are almost identical, with greater than 99·9% sequence identity, suggesting that SARS‐CoV‐2 has originated from one source, from Wuhan and has spread relatively rapidly worldwide, within a short period. Through the novel furin cleavage site in its S protein, the SARS-CoV-2 utilizes and affects ACE in multiple ways. Apart from its entry through the ACE2, SARS-CoV-2 subsequently down-regulates ACE2 expression resulting in unopposed angiotensin II accumulation and local RAAS activation, which deranges homeostasis and exacerbates the inflammation and potentiates the tissue injury in lungs and other organs. In this perspective and in light of the studies and available data, it is advisable that an ACE inhibitor or ARB should be continued, rather than withdrawn in favour of another antihypertensive, in known or likely Covid-19 patients with stable condition.
Evolving Covid-19 Therapeutics
Presently there are no effective drugs for treatment of the Covid-19. The integrative network-based systems pharmacology methods have been used for rapid identification of repurposable drugs and drug combinations for the potential treatment. While new and repurposed drugs, including antiviral agents are being tested in various clinical trials, some of the promising drugs are simultaneously being recruited off-label for compassionate use or as experimental drugs to treat in desperate situations and otherwise dying patients. There has been a growing interest for the use of chloroquine and hydroxychloroquine as potential treatment in the interim till a specific treatment is available. There have been identified certain proteins in the host immune pathways which can be targeted for blocking viral replication by potential drugs or antibodies. Alternatively, the passive antibody transfer from pooled convalescent patient sera may seem another option. As obvious, not all clinical trials will be successful, but having so many efforts in the direction, some may succeed and provide plausible solutions.
Keywords: ACE2 Expression; Chloroquine and Hydroxychloroquine; Covid-19; Furin Cleavage site, Horseshoe Bats; Malayan Pangolin; Pooled Convalescent Sera; Repurposed Drugs; S Protein; SARS-CoV-2; Wuhan Epidemic
Infectivity and Pathogenesis
The Agent Factors
Origin and Evolution of 2019-nCoV: The Coronaviruses (CoVs) belong to the subfamily Coronavirinae in the family of Coronaviridae of the order Nidovirales. The CoVs are enveloped RNA viruses, having the genome composed of a single‐stranded positive‐sense RNA (+ssRNA), ~30kb with 5′‐cap structure and 3′‐poly‐A tail. The large CoV genome is related to the special features of the CoV RTC, which contains several RNA processing enzymes and encodes for several structural proteins (sps) and non-structural proteins (nsps) [1]. The sps having a critical role in viral RNA transcription and synthesis, are referred to as replicasetranscriptase proteins, whereas the nsps, also referred to as nichespecific proteins, though nonessential for virus replication but confer selective advantages for survival and tissue invasion [2]. The CoVs are further divided into four lineages, A to D. The lineage B includes the severe acute respiratory syndrome (SARS)-CoV and the novel SARS-CoV-2, whereas the lineage C includes Middle East respiratory syndrome (MERS)-related CoV. The six human CoVs were identified till last year and included HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS-CoV and MERS-CoV. The SARSCoV- 2 or 2019-nCoV is the latest and most virulent member of the CoV family that can infect humans [3].
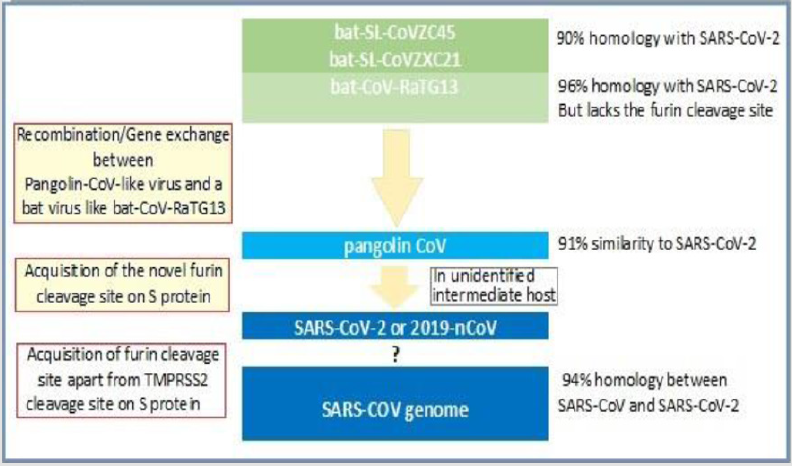
The SARS-CoV-2 shares a high sequence identity to SARS-CoV. The protein sequence analyses have documented that the amino acid similarity of the conserved nsps between SARS-CoV-2 and SARS-CoV is as high as 94.6%, suggesting that they are closely related. The homology between the SARS-CoV-2 genome and CoV bat viruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21 is approximately 90 percent, whereas it is approximately 96% with the bat SARS-like CoV (bat-CoV-RaTG13) genome [4]. There is consensus among the scientific community that the SARS-CoV-2 originated in horseshoe bats. But, the bat-SL-CoVZC45 and bat-SL-CoVZXC21 are not direct ancestors of 2019-nCoV. Instead the bat CoV may infected another animal, an intermediate host, which subsequently transmitted the virus to humans (Figure 1). The isolation of a CoV in the Malayan pangolins with high similarity to SARS-CoV-2 links these animals as being the potential intermediate host [5].
The genetic sequence of CoV, discovered in the lung samples of
Malayan pangolins has a ~91% similarity to SARS-CoV-2. In addition,
the pangolin CoV exhibits 100%, 98.2%, 96.7% and 90.4% amino
acid identity with the SARS-CoV-2 E, M, N and S genes, respectively.
The ancestor bat CoVs, have a 19 amino acids dissimilarity,
whereas the pangolin CoV has only 5 amino acids dissimilarity
for the S protein from SARS-CoV-2. In particular, the receptorbinding
domain of the S protein of the Pangolin-CoV is virtually
identical to that of SARS-CoV-2, with one amino acid difference. The
comparison of available genomes appears to suggest that SARSCoV-
2 might have originated from the recombination of a Pangolin-
CoV-like virus with a virus similar to bat-CoV-RaTG13, which has a
striking 96% similarity to the SARS‐CoV‐2 virus but lacks the furin
cleavage site. Further, the infected pangolins show clinical signs
and histopathological features akin to SARS-CoV-2 in experimental
models and harbour the antibodies which react with the S protein
of SARS-CoV-2 [6]. The genetic sequence analysis of the bat CoVs,
pangolin CoV and SARS-CoV-2 appears to link the CoVs, suggesting
that viruses from the bats and pangolin may have exchanged the
genes at some point before spilling and infecting human-beings [7].
The pangolins are, thus, the most likely intermediate host, though
other potential intermediate host(s), cannot be ruled out.
The pangolin CoV lacks a characteristic trait seen in SARSCoV- 2, that has helped it to leap to humans, the adaptation may have been acquired in another, yet unidentified, animal before the Wuhan epidemic spreading around the globe [8]. There has been identified a unique peptide (PRRA) insertion in the human SARS‐CoV‐2 virus in the S protein between its S1 and S2 subunits, which seems to be involved in the proteolytic cleavage of the spike protein by cellular furin-like proteases, which are widely expressed in a variety of organs including lungs and respiratory tracts, gastrointestinal tract, liver, pancreas and brain [9]. This furin cleavage site is distinct from SARS-CoV and other CoVs which only contain a trypsin or TMPRSS2 cleavage site. With the novel furin cleavage site on the S protein, four amino acid residue insertion at the boundary between the S1 and S2 subunits, the SARS-CoV-2 appears to gain a potentially high infectivity and transmissibility as compared to SARS-CoV, due to the near-ubiquitous distribution of furin-like proteases in various tissues [10]. The sequences of SARSCoV‐ 2 from various patients from different countries are almost identical, with greater than 99·9% sequence identity. As a typical RNA virus, the average evolutionary rate for CoVs is approximately 10-4 nucleotide substitutions per site per year, with mutations arising during every replication cycle. This finding suggests that SARS‐CoV‐2 has originated from one source, from Wuhan and has spread relatively rapidly worldwide, within a short period [11].
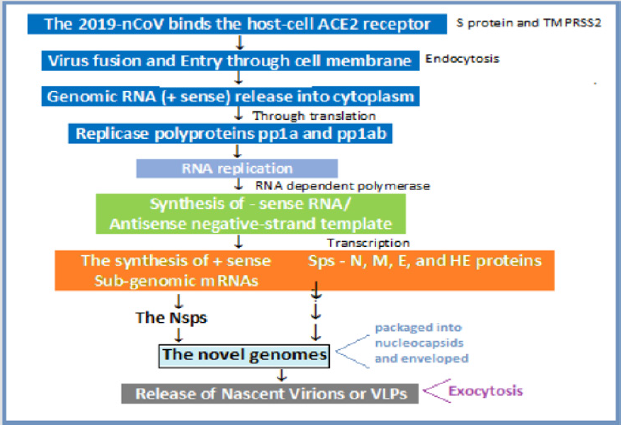
The CoV Transcription and Expression: The CoVs have complex gene expression and replication cycle, encompassing ribosome frameshifting for genome translation, the synthesis of both genomic and sub-genomic components, and the assembly of progeny virions. Further, the CoV transcription involves the production of multiple sub-genomic mRNAs that contain sequences corresponding to both ends of the genome through a process of discontinuous transcription [12].The expression of the CoV replicase-transcriptase (RT) protein genes is mediated by the translation of the genomic RNA. Whereas, the sub-genomic messenger RNAs (mRNAs) possess common 5′‐leader and 3′‐ terminal sequences. The genome and sub-genomes of a typical CoV contain at least six open reading frames (ORFs). The first ORFs (ORF1a/b), comprising of about two‐thirds of the whole genome length, produce two polypeptides, pp1a and pp1ab, which are processed by virally encoded protease into 16 nsps, 1 to 16. The other ORFs on the rest of one‐third of the genome near the 3′‐ terminus encode 4 main sps - spike (S), membrane (M), envelope (E) and nucleocapsid (N) proteins. Besides the sps, accessory proteins, such as HE protein, 3a/b protein and 4a/b protein are also encoded. The sps and accessory proteins are translated from the sub-genomic RNAs (sgRNAs) [13].
The 2019-nCoV Structural Components: Spike (S) Protein: The glycosylated S protein is a fusion protein and mediates attachment of the virus to the host cell surface receptors and subsequently facilitate its entry through the cell membrane into the host cell. The S protein has two subunits: the globular S1 subunit is involved in receptor recognition and has a large receptor-binding domain (RBD) structure organized in four distinct domains, whereas the S2 subunit forms the stalk of the spike molecule and contains the key protein segments including the fusion peptide, two heptad-repeat regions (HR1 and HR2) that facilitate viruscell fusion, and has been conserved across different genera of CoV species [14]. There are various receptors that bind to different CoVs, such as ACE2 for SARS-CoV29 and CD26 for MERS-CoV. There being a structural similarity between the RBDs of SARS-CoV and SARS-CoV-2, SRS-CoV-2 also uses ACE2 as its receptor, despite the presence of amino acid mutations in its S1 RBD [15].
The CoV S glycoprotein has N-terminal region (NTR, domain A) and the C-terminal region of S1 (CTR, domains B, C, and D) which bind to the host ACE2 receptors and function as RBDs [16]. The major difference in S1 subunit of SARS-CoV-2 as compared to SARS-CoV, is the 3 short insertions its N-terminal domain, which confer the sialic acid binding activity and 4 out of 5 key residues changes in the receptor-binding motif [17]. Other Major Structural Proteins: In addition, the CoV genome encodes other major sps, such as, nucleocapsid (N) protein, membrane (M) protein and the envelope (E) protein. The N protein is the most abundant protein in CoV and binds to the CoV RNA genome, leading to formation of the helical nucleocapsid. It is composed of two domains, an N-terminal domain (NTD) and a C-terminal domain (CTD). The genomic packaging signal binds specifically to the C-terminal RNA binding domain, whereas NTD binds to nsp3, a key component of the replicase complex and the M protein. The N protein is localised to the endoplasmic reticulum (ER)-Golgi region and involved in nascent virion assembly and budding. Its high hydrophilicity has been related to the host cellular response and potent immunity following the viral infection.The M protein, another major structural protein, helps in defining the shape of viral envelope. It is the central organiser of CoV assembly and interacts with all other major coronaviral sps. Interaction of S with M is necessary for retention of S in the ER-Golgi intermediate compartment (ERGIC)/ Golgi complex and its incorporation into new virions. The binding of M to N stabilises the nucleocapsid (N protein-RNA complex) core.
The E protein is abundantly expressed inside the infected cell and localised at the ER, Golgi and ERGIC, and during the replication cycle a small portion of E protein is incorporated into the virion envelope along with a major contribution by M protein [18]. The E protein appears to be involved at multiple stages of the replication cycle, from assembly and induction of membrane curvature to budding and release, and inflammation, apoptosis and autophagy. The deletion of E from SARS-CoV attenuates the virus. The hemagglutinin-esterase (HE) protein is attached to the viral surface. It contains a lectin-binding domain that mediates binding to O-acetylated sialic acids. It possesses sialate-O-acetylesterase receptor-destroying enzyme activity, though which viral attachment to non-permissive cells is prevented. The HE protein appears to facilitate virus entry into target cells after binding to the main entry receptor.
Receptor Tethering, Viral Invasion and Replication: The Viral Tethering and Cell Entry: Following receptor binding, the SRS-CoV-2 gains access to the host cell cytosol, accomplished by acid-dependent proteolytic cleavage of S protein by a furin like protease. The S protein cleavage occurs at two sites within the S2 portion of the protein, the first cleavage separates the RBD and fusion domains of the S protein and the second cleavage exposes the fusion peptide which inserts into the cell membrane, followed by joining of two heptad repeats, HR1 and HR2, in S2 forming an antiparallel six-helix bundle. The formation of the bundle allows the blending of viral and cellular membranes, resulting in fusion and ultimately release of the viral genome into the cytoplasm. Replicase Protein Translation and Expression: The next step is the translation of the replicase gene from the viral genomic RNA. Viral RNA synthesis follows the translation and assembly of the viral replicase complexes and generation of genomic and sub-genomic RNAs. The replicase gene encodes two large ORFs, rep1a and rep1b, which express co-terminal polyproteins (pps), pp1a and pp1ab. In order to express the pps, the virus utilizes an RNA pseudoknot that cause ribosomal frameshifting from the rep1a into the rep1b ORF. The pps are subsequently cleaved into the individual nsps by proteases encoded by the virus. Various nsps assemble into the replicasetranscriptase complex (RTC) for RNA replication and transcription of the sub-genomic RNAs. The sub-genomic RNAs serve as mRNAs to generate the structural and accessory proteins (Figure 2).
Functions of CoV non-structural proteins: The nsps contain
domains to carry out various genomic as well as supportive
functions. The nsps promotes cellular mRNA degradation (nsp1)
and blocks host cell innate immune response (nsp1 nsp3), cleave pps
(nsp5), bind to prohibitin proteins (nsp2), interact with N protein
(nsp3), act as processivity clamps for RNA polymerase (nsp7 and
8) and transmembrane scaffold proteins (nsp4 and 6). In addition,
they promote cytokine expression (nsp3), stimulates viral 3′-5′
exoribonuclease (nsp10 and 14) and N7 methyltransferase (nsp10
and 14), shield viral mRNA from recognition (nsp16) and bind viral
RNA (nsp9). They also modulate the enzymes RNA-dependent RNA
polymerase (nsp12), RNA helicase 5’triphosphatase (nsp13) and
viral endoribonuclease (nsp15), and act as cofactor for nsp14 and
16 (nsp10).
Virion Assembly and Release: Following replication and subgenomic
RNA synthesis, the viral sps, S, N, M, E and HE are translated
and inserted into the endoplasmic reticulum (ER). The sps move
along the secretory pathway into the ERGIC. The viral genomes are
encapsidated by the N protein bud into membranes of the ERGIC
containing viral structural proteins, forming nascent virions or
virus like particles (VLPs). The M protein then directs most proteinprotein
interactions required for assembly of virions and expressed
along with E protein to produce virion envelope. The N protein
enhances VLPs formation and the S protein is incorporated into
virions at this step. The M protein also binds to the nucleocapsid,
promoting the completion of virion assembly. Following assembly,
VLPs are transported to the cell surface in vesicles and released by
exocytosis.
The Host Factors
Angiotensin-Converting Enzyme 2 (ACE2): The ACE2, also known as ACEH (ACE homolog), is an integral membrane protein and a zinc metalloprotease of the ACE family. The human ACE2 protein sequence consists of 805 amino acids, including a N-terminal signal peptide, a single catalytic domain, a C-terminal membrane anchor and a short cytoplasmic tail. ACE2 cleaves angiotensin I and II as a carboxypeptidase. ACE2 is located as an ectoenzyme on the surface of endothelial and other cells. There is an abundant presence of ACE2 in nasal mucosa and nasopharynx, oral mucosa and tongue, airways and lung alveolar epithelial cells. There is rich ACE2 surface expression on arterial and venous endothelial cells, and arterial smooth muscle cells. The abundant ACE2 expression has been linked with the pathogenesis involving vasculitis, deranged immune function, extensive pulmonary inflammation, and diffuse alveolar damage with hyaline membrane formation, and the severe clinical manifestations [19]. The oral cavity is a potentially high-risk route for SARS-CoV-2 infection and should be a part of future prevention strategy in dental clinical practice as well as daily life [20]. Apart from this, there is a high ACE2 expression in oesophageal upper and stratified epithelial cells, absorptive enterocytes from ileum and colon, cholangiocytes, myocardial cells, renal proximal tubule cells and bladder urothelial cells, skin, lymph nodes and brain. The widespread ACE2 tissue distribution in various organs may explain the multi-organ dysfunction observed Covid-19 in patients.
The ACE2 is an important regulator of the renin-angiotensin
system (RAS). Angiotensin I, having no direct biological activity,
exists as a precursor to angiotensin II, and converted to the latter
through removal of two C-terminal residues by the ACE2 primarily
in the lung but also in kidneys, endothelial cells, and brain.
Angiotensin II acts on venous and arterial smooth muscle to cause
vasoconstriction and increases vasopressin production in the CNS.
It also stimulates aldosterone secretion. It appears that apart from
gaining its entry through the ACE2, the SARS-CoV-2 subsequently
down-regulates the ACE2 expression leading to loss of its protective
effects in various organs, which may have significant impact on the
pathogenesis of the disease.
There is reduced ACE2 expression with aging in both genders.
It is slightly reduced in young-adult and middle-aged groups but
reduced significantly in the elderly age group. In animal studies,
though there was no gender-related difference of ACE2 expression
in adult mice, a higher ACE2 content was noticed in old female
than male [21]. The epidemiological studies have documented that
different sex and age groups have different susceptibility to SARSCoV-
2 infection, which may be linked to ACE2 expression. There is
a skewed relationship with various host factors and the severity
and mortality of the disease, with the male sex, elderly population
and those with coexisting chronic diseases being the most affected.
Further, as per the preliminary epidemiological data, there is an
exponential increase in disease severity and mortality in those
beyond the sixth decade of life with cardiovascular disease and
diabetes [22]. The exaggerated proinflammatory profile is appears
to be a salient feature in those suffering from hypertension, heart
disease and diabetes. In addition, the elderly, especially those with
hypertension and diabetes, have reduced ACE2 expression and
upregulation of angiotensin II proinflammatory signaling. The
SARS-CoV-2 binding to ACE2 may exaggerates this proinflammatory
milieu, predisposing these population groups to greater disease
severity and mortality.
S Protein-ACE2-Protease Host Cell Entry: The ACE2, expressed on the surface of various tissues including airway epithelial cells, binds with the S protein. The RBD on the spike, mediates the interaction with the host-cell receptor, ACE2 for both SARS-CoV and SARS-CoV-2. The spike RBD is capable of folding independent of the rest of the spike protein and contains the structural information for host receptor binding. The host protease processing during viral entry is a may be a significant barrier infection, but has been overcome by hCoV including SARSCoV-2 to the agent’s advantage and after binding the receptor, a host protease cleaves the spike, releasing the spike fusion peptide and facilitates the virus entry [23]. The CoV entry into the host cell is a multi-step process. It involves several distinct domains in spike mediating the virus attachment to the cell surface, receptor engagement, protease processing and membrane fusion. After bonding with the host receptor, the host-cell protease cleaves spike and releases the fusion peptide, allowing for host-cell entry. Thus, armed with the SARS-CoV spike, the SARS-CoV-2 is capable of using human ACE2 efficiently, which may account for the human-tohuman transmissibility of the virus [24].
The ACE2 is predominantly expressed in surfactant-secreting
type II alveolar cells in lungs, bronchiolar epithelium, and
endothelium and smooth muscle cells of pulmonary vessels.
Further, it is more abundantly expressed in the apical than the
basolateral areas of lungs. Furthermore, the ACE2 expression is
proportionately correlated to the epithelial differentiation of the
alveolar tissue. The undifferentiated cells poorly express ACE2,
while well-differentiated cells richly express ACE2. The studies
indicate that infection of human airway epithelia by SARS-CoVs
correlates with the state of cell differentiation and ACE2 expression
and localization. These findings have implications for understanding
disease pathogenesis associated with the SARS-CoV infections [25].
The lungs appear to be the most vulnerable target organs
for SARS-CoV and SARS-CoV-2 because of the vast surface area,
making the lung highly susceptible to inhaled viruses. About 83
percent of ACE2-expressing cells are alveolar type II cells, which
are exposed and can serve as a reservoir for the viral invasion. In
addition, the ACE2-expressing alveolar type II cells contain high
levels of viral process-related genes, including regulatory genes
for viral processes, viral life cycle, viral assembly, and viral genome
replication. Further, it has been claimed that ACE2 is not only the
entry receptor of the virus but also protects from lung injury and
high lethality associated with SARS-CoVs infection could be because
of dysregulation of the pulmonary protective mechanisms [26].
Pathogenesis of CoV Infections: Coronaviruses cause a large variety of severe diseases in livestock and other animals such as pigs, cows, chickens, dogs and cats, which has led to significant research on these viruses. For instance, Transmissible Gastroenteritis Virus (TGEV) and Porcine Epidemic Diarrhoea Virus (PEDV) cause severe gastroenteritis in young piglets, leading to significant morbidity and mortality, and economic loss. The SARS-CoV, a 2b β-coronavirus, was identified as the causative agent of the Severe Acute Respiratory Syndrome (SARS) outbreak that occurred in 2002– 2003 in the Guangdong Province of China, leading to approximately 8098 cases and 774 deaths, amounting to a mortality rate of 9%. The rate was much higher in elderly individuals, with mortality rates approaching 50% in individuals over 60 years of age [27]. It appears that the SARS-CoV originated in bats as a large number of Chinese horseshoe bats harbour sequences of SARS-related CoVs and serological evidence for a prior infection with a related CoV. In fact, two novel bat SARS-related CoVs, bat-SL-CoVZC45 and bat- SL-CoVZXC21, were identified as having significant similarity to SARS-CoV. They were also found to use the same receptor, ACE2. Transmission of SARS-CoV was relatively inefficient, as it only spread through direct contact with infected individuals after the onset of illness, and the outbreak was controllable through strict quarantine.
Another novel human CoV, named Middle East Respiratory Syndrome-CoV (MERS-CoV), was found to be the causative agent in a series of highly pathogenic respiratory tract infections in Saudi Arabia and other countries in the Middle East, which emerged in the Middle East in 2012. It was thought that camel-to-human spill over contributed to the outbreak. The outbreak did not accelerate later in 2013, though sporadic cases continued throughout the rest of the year. From 2012 through May 31, 2019, MERS-CoV has infected 2,442 persons and killed 842 worldwide amounting to case fatality rate of ~29 percent [28]. The MERS-CoV utilizes Dipeptidyl peptidase 4 (DPP4) as its receptor in humans and certain species such as bats, camels, rabbits and horses to establish infection. The virus is unable to infect mouse cells due to difference in the structure of DPP4. Owing to the lack of effective therapeutics or vaccines, the best measures to control hCoVs, remain an efficient public health surveillance system coupled with rapid diagnostic testing, and isolation and quarantine. The past and likely future emergence of pathogenic zoonotic CoVs, the gross social and economic impact of hCoVs infections and the lack of effective antiviral strategies make it obvious that our preparedness to prevent and treat CoV infections is limited [29]. This highlights the importance of advancing our knowledge on the emergence, infectivity, replication and pathogenesis of the CoVs.
The Host Cell Immune Response: In general, the host cells
respond to the virus infection by recruiting an innate antiviral
response to limit the spread of the infection and resorting to induce
an adaptive immune response to eventually clear the virus. In the
case of CoVs and other +RNA viruses, the innate immune system
is triggered by recognizing dsRNA and 5′-triphosphate-bearing
RNA molecules arising as replication intermediates in the cytosol
by the intracellular sensors of the Rig-I-like receptor (RLR) family,
such as retinoic acid-inducible gene 1 (RIG-I)[30] and melanoma
differentiation-associated protein 5 (MDA-5) which are expressed
in various host cells [31]. The infections are recognized by the
cytosolic sensors RIG-1, Mda5 and stimulator of interferon genes
(STING) and result in the expression of IFN-β and an inflammasome
response. The RLRs, RIG-1, Mda5 and LGP2 recognize the viral
ligands. Mda5 is one of the pattern recognition receptors (PRRs)
that recognize cytoplasmic viral ligands [32].
For recognition of CoV RNAs, MDA-5 seems the most important
cytosolic sensor. The toll-like receptors (TLRs) which are expressed
on the cell surface or reside in the endosomes of immune cells can also recognize CoVs nucleic acids or proteins. Activation of one
or more of these sensors generally leads to the activation of the
transcription factors IFN-regulatory factor 3 and 7 (IRF3, IRF7)
and NF-κB, which stimulate the expression and secretion of Type-I
IFN and pro-inflammatory cytokines. Following this, the JAK-STAT
(Janus kinase/signal transducers and activators of transcription)
signaling cascade is activated leading to expression of various
antiviral interferon-stimulated genes (ISGs) resulting in an antiviral
state of the infected cells.
The CoV infection and stages of replication are associated with
the endoplasmic reticulum (ER) stress. The ER can sustain a high
load of protein content without being overwhelmed. However,
when the ER’s capacity for folding and processing proteins is
exceeded, unfolded or misfolded proteins rapidly accumulate in
the lumen and the ER stress response is induced. The ER stress
leads to activation of Unfolded Protein Response (UPR) pathways
through the induction of protein kinase RNA-like endoplasmic
reticulum kinase (PERK). The activated UPR leads to elevated level
of phosphorylated Eukaryotic Initiation Factor 2 alpha (eIF2α)
resulting in the promotion of a pro-adaptive signaling pathway by
the inhibition of global protein synthesis and selective translation
of Activating Transcription Factor 4 (ATF4). In addition, during
conditions of prolonged ER stress, several pro-apoptotic genes
are induced, and there is failure of synthesis of anti-apoptotic Bcl-
2 proteins. The ER stress-mediated leakage of calcium into the
cytoplasm also leads to the activation of apoptosis effectors. These
measures eventually trigger apoptosis, which is utilized by the host
cells to inhibit viral replication. PKR (serine/threonine protein
kinase) is a key player in the innate immune response to RNA virus
infection by upregulating antiviral gene expression and enhancing
synthesis of interferons (IFNs).
The CoVs Countermeasures: The viruses, on the other hand,
have evolved strategies to suppress and overcome the immune
responses, which influence the pathogenesis, course of the disease
and persistence the virus in the host. The CoVs strategically
counteract PKR-mediated signaling to prevent the translational
shut-off due to eIF2α phosphorylation. In case of the MERS-CoV,
the ORF4a protein counteracts the PKR-induced formation of
stress granules, probably by binding viral dsRNA to shield it from
detection by PKR. The S proteins of both SARS-CoV and SARS-CoV-2
also interact with eIF3F, to modulate host translation, including
the expression of the pro-inflammatory cytokines, and IL-6 and 8
at a later stage of infection. These interactions play an important
regulatory role in CoV pathogenesis, and disease course and
prognosis [33].
Apart from modulating eIF2α phosphorylation, the hCoV
manipulates the translation machinery through nsps. The nsp1
protein, an inhibitor at multiple steps of translation, inhibits
48S initiation complex formation and its conversion into the 80S
initiation complex, apart from directly binding to the 40S ribosomal subunit to stall translation. Further, the nsp1 and 40S complex
induce cleavage of cellular mRNAs to suppress the translation.
The viral nsps, especially nsp3, nsp4 and nsp6, appear to drive
the formation of replication structures such as double-membrane
vesicles (DMVs), tubules, zippered ER and convoluted membranes
forming a reticulovesicular network in the cytosol of infected cells.
In addition, various RNA-binding proteins in the infected host cells
fail to interact and block the CoV 5′ UTR, the 3′ UTR and poly(A)-
tail [34]. In addition, the CoV nsp15 inhibits retinoblastoma protein
(pRb), a tumor suppressor protein, resulting in enhanced expression
of genes normally repressed by pRb [35]. The overexpression
of the SARS-CoV 3a protein leads to G1 arrest and inhibition of
cell proliferation. Apart from this, SARS-CoV infection decreases
p53 expression related to antiviral effect and able to enhance its
replication in the cells lacking p53 depleted cells [36].
The Protein–Protein Interactions (PPIs): The PPIs can be
host–host, virus–virus or virus–host PPIs. Through PIPs the viral
proteins establish interactions with host proteins. The viral protein
domains are basic units for viral–host protein interactions and the
mutations at protein interfaces can reduce or increase the binding
affinities. During the course of the viral invasion and infection,
both the agent and cellular proteins are constantly competing for
bonding. The protein–protein interactions (PPIs) in virus–host
systems are mediated by domain–domain interactions (DDIs),
involving molecular recognition via amino acid residues located at
interfaces of interacting domains [37]. Apart from this, endogenous
interfaces mediating viral–viral or host–host interactions, are
constantly targeted and inhibited by exogenous interfaces
mediating the interactions. In case of viral infection, the virus–host
PIPs, the interacting proteins are constantly losing and regaining
their binding sites in order to evade or optimize interspecific PPIs.
The exogenous interfaces lend competition for such molecular
reactions and interfere with host–host protein interactions leading
to alterations in cellular metabolism [38].
The host factors evolve to retain or restore their recognition
capabilities to bind and neutralize viral agent. Whereas, through
virus–host interactions, the viral proteins tend to target more central
and highly connected host proteins and maintain and adapt their
capabilities via molecular evolutions and, including conservation,
HGT, gene duplication and molecular mimicry [39]. Horizontal gene
transfer (HGT) is a process of genome recombination by means
of which the virus acquires genes from non-parental organisms,
optimizes and integrates into its virus–host network. Through
gene duplication, the virus acquires duplicated genes in the viral
genomes to become more lethal. As the virus evolves at faster
mutation rate, it can rapidly acquire new binding partners by
mimicking and targeting interfaces of host proteins.
Immune Response/Inflammasome Activation: There is a complexity of the virus–host interactions that occur within the cell, tissues or the whole body. The SARS-CoV-infected peripheral blood mononuclear cells lead to the upregulation of expression of various cytokines, including IL-8 and IL-17, and the activation of macrophages and the coagulation pathways. The protein kinases, the key regulators in signal transduction, control a wide variety of cellular processes. They are linked to cellular immune responses, like interleukin (IL) signaling, IL-6 and -8, and influence the CoV infection and CoV-induced inflammation. There occurs downregulation of a large number of mRNAs, including those encoding proteins involved in translation, leading to the host translational shut-off in CoV-infected cells due to a stress response and concomitant mRNA decay. In addition, the heterogeneous nuclear ribonucleoproteins (hnRNPs) influence the maturation of nascent nuclear RNAs into messenger RNAs (mRNAs) and stabilize their cellular transport and control their translation. In general, the difference in the immune and inflammatory responses determines the outcome of the infection and responsible for the differential activation of the Signal transducer and activator of transcription 3 (STAT3) pathway, involved in lung inflammation and cellular repair [40].
The CoV nucleocapsid (N) protein plays a multifunctional role in the virus life cycle, from regulation of replication and transcription to genome packaging and release of VLPs to modulation of host cell defence processes. In addition, various viral proteins interact with host ribosomal and nucleolar proteins, helicases and the hnRNPs [41]. The viruses encode proteins to modulate the cellular factors to modify the immune response pathways to avoid their detection and exploit the extensive network of the host cell’s signalling pathways to promote their replication and propagation. The inflammasome activation by CoV E was first reported in porcine reproductive and respiratory syndrome virus (PRRSV), when it documented that PRRSV-encoded small envelope protein E, an ion channel-like protein, triggered the activation of inflammasomes. Blocking ion channel activity with amantadine significantly inhibited activation of the inflammasome [42]. Recently, the transport of Ca2+ by SARSCoV E has been shown to trigger inflammasome activation [43]. Following which, there has been shown ER stress and apoptosis to occur, and the inflammatory lung damage in SARS-CoV-infected mice. Inhibiting the CoV E ion channel activity limited the CoV E viroporin induced CoV pathogenicity. Autophagy is a reparative cellular process that recycles excess or damaged cellular material inside the cell ensure its survival. The RNA viruses, including CoVs, appear to exploit autophagy for the purpose of viral replication and propagation.
Immunological Basis of Severe Covid-19: In the severely
ill SARS-CoV-2 patients, the disease may progress rapidly from
viral pneumonia to acute respiratory failure. The neutrophil-tolymphocyte
ratio (NLR) has been identified as an independent risk
factor for severe illness in these patients. The patients with age ≥ 50
and NLR ≥ 3.13 denote a severe illness and should have rapid access
to intensive care unit and respiratory support [44]. The pyroptosis,
a form of inflammatory form of programmed cell death and appears
to be the possible mechanism for the increased virulence of SARS CoV-2[45]. The SARS-CoV Viroporin 3a triggers the activation of the
NLRP3 inflammasome and the secretion of IL-1β by macrophages,
suggesting SARS-CoV induced cell pyroptosis [46]. In addition,
the patients infected with SARS-CoV-2 have increased IL-1β in
the serum, an indicator of the pyroptosis [47]. Further, the rising
IL-1β suggests activation of cell pyroptotic activity. The severe
SARS-CoV-2 infection is likely to cause cell pyroptosis, especially in
lymphocytes, through the activation of NLRP3 inflammasome.
About thirty percent of patients demonstrated neurological
manifestations out of a sample of 214 newly diagnosed Covid-19
patients [48]. The central nervous system (CNS) involvement is
indicated by headache, dizziness, disturbance of consciousness,
acute cerebrovascular disease including epilepsy, and peripheral
nervous system symptoms such as decreased taste, decreased smell,
and appetite. The SARS-CoV-2 appears to invade the CNS through
blood or retrograde neuronal circuits, leading to neurological
damage and viral encephalitis.
Evolving COVID-19 Therapeutics
Outlining Solutions for Treatment
Presently there are no effective drugs for treatment of the Covid-19, which is now pandemic with millions of confirmed cases worldwide. While new and repurposed drugs are being tested in clinical trials, some of the promising drugs are simultaneously being recruited off-label for compassionate use or as experimental drugs to treat in desperate situations and otherwise dying patients. As obvious, not all clinical trials will be successful, but having so many efforts in progress, some may succeed and provide plausible solutions. Right now, though, there prevail confusion and despair. The future seems afflicted with dormant therapeutic options as well as faux Espoir or false hopes. See my review article – Preprint – “Exploring Pathophysiology of Covid-19 Infection: Faux Espoir and Dormant Therapeutic Options”. On www.researchgate.net DOI: 10.13140/RG.2.2.16995.09764. please visit- https:// www.researchgate.net/publication/340175439._Exploring_ Pathophysiology_of_COVID-19 Infection: Faux_Espoir_and_ Dormant_Therapeutic_Options.
>Understanding the PPIs offers an opportunity to target both viral-host and intraviral interactions to halt the viral replication and propagation. The virus binding and entry are the first steps of the replication cycle that can be targeted with inhibitors. There are inhibitors of endosomal acidification, such as ammonium chloride, chloroquine and hydroxychloroquine, which appear to block the entry of CoVs including SARS-CoV and SARS-CoV-2. In addition, peptides have been explored which can block recognition and fusion by interfering with the interaction between the HR1 and HR2 domains of the S protein. Apart from drugs acting as inhibitors directed at viral components, there are evolving therapeutic strategies involving host-directed approaches, based on the pathogenesis and virus–host interactions. The host-directed approach can yield a broad-spectrum therapeutic strategy and may lower the chance of development of antiviral resistance. The interferon (IFN) has been shown to trigger the innate immune response in CoV-infected cells, leading to transcription of various interferon stimulated genes (ISGs) that may play a role in controlling and eradicating the infection [49].
Stalling ACE2-mediated COVID-19 Entry
Spike Protein-Based Vaccine: Development of a spike-1 subunit protein-based vaccine relies on the fact that ACE2 is the SARS-CoV-2 receptor [50]. The subunit vaccines for SARS-CoV may work by eliciting an immune response against the S-spike protein to prevent its docking with the host ACE2 receptor. In this context, the Novavax Inc, headquartered in Maryland, has developed immunogenic virus-like nanoparticles based on recombinant expression of the S-protein [51]. The Clover Biopharmaceuticals is developing a subunit vaccine consisted of a trimerized SARSCoV- 2 S-protein using their patented Trimer-Tag technology [52]. Another subunit vaccine being developed and tested is comprised of the receptor-binding domain (RBD) of the SARS-CoV S-protein [53]. Initial findings have documented that the SARS-CoV and SARSCoV- 2 RBDs exhibit more than 80% amino acid similarity and bind to the same ACE2 receptor offer an opportunity to develop either protein as a subunit vaccine.
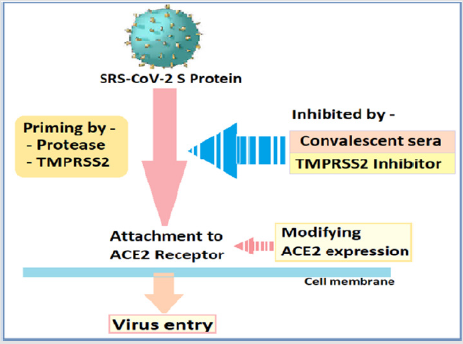
Inhibition of TMPRSS2 Activity: The S protein of CoVs facilitates viral entry into target cells through attachment with ACE2 receptors. The priming of S protein by protease, namely, transmembrane protease serine 2 (TMPRSS2) is a prerequisite for S protein cleavage at the S1/S2, which frees S2 subunit essential for entry and translocation of the SARS-CoV-2 genome through interaction with the ACE2 receptor (Figure 3).The protease, TMPRSS2 is a potential target for therapeutic intervention and the serine protease inhibitor, camostat mesylate, has been shown to block TMPRSS2 activity and to prevent SARS-CoV-2 entry into the host cells [54] In the animal study, Camostat protected 6 out of ten mice from lethal infection with SARS-CoV. The results, thus, indicate that camostat or similar serine protease inhibitors may be a potential treatment option for SARS-CoV and SARS-CoV-2 infections [55]. The drug, camostat mesylate is already approved in Japan for the treatment of a number of non-infectious conditions.
Convalescent SARS Sera: The convalescent SARS patients exhibit a neutralizing antibody response directed against the S protein, which can be detectable as long as 24 months or more following the infection56. It has been found that the antibodies found in the recovering SARS patients sera cross-neutralized SARS- 2-S-driven entry as well, as apparent from being able to hamper the SARS-CoV-2 S protein-driven entry into Vero-E6 cells, in vitro [56]. The finding is important as it reveals an important commonality between SARS-CoV-2 and SARS-CoV infection and a potential therapeutic intervention.
Modifying ACE2 Expression
The interaction between ACE2 and SARS-CoV is a therapeutic target. Theoretically, the anti-ACE-2 antibodies can block SARSCoV- 2 binding to the receptor, disrupt the interaction, prevent the virion entry and the alveolar cell damage. The treatment with anti- ACE-2 antibodies, thus, disrupts the interaction between virus and receptor. The same results can be achieved through enhancing the ACE2 expression by treatment with soluble ACE2.
It has been demonstrated in mice experiments that SARSCoV downregulates ACE2 protein by binding its spike protein, contributing to severe lung injury [57]. It appears that, ACE2 competitively binds with SARS-CoV-2 not only to neutralize the virus but also rescue cellular ACE2 activity which negatively regulates the renin-angiotensin system (RAS) to protect the lung from injury [58]. Apart from this, angiotensin-(1-7) is generated by degradation of Ang II by ACE2 has vasoprotective functions. The enhanced ACE activity and decreased ACE2 availability may, thus, contribute to oxidative stress and subsequent lung injury. ACE2 deficiency may be critical in a variety of co-existing disease states, including hypertension, diabetes, renal impairment and cardiovascular disease, and in elderly with physiological reduced ACE2 availability.
Functionally, there are two forms of ACE2. The membranebound ACE2 contains a transmembrane domain and an extracellular domain, which is used by SARS-CoV-2 as the receptor. The other, soluble form of ACE2 lacks the membrane anchor and circulates in small amounts in the blood. In this context, treatment with a soluble form of ACE2 may slow the viral entry into host cells, as the soluble ACE2 acts as a competitive interceptor of SARS-CoV by preventing its binding to the membrane-bound ACE2, limits cell entry and spread, and protects the lungs from injury. The recombinant human ACE2 (rhACE2; APN01, GSK2586881) has been found to be safe in healthy volunteers and in a small cohort of patients with ARDS [59].
The Stapled Peptides
Therapies using small-molecule drugs, allows the drugs to cross cell membranes but may lack selectivity. Further, the smallmolecule drugs may not be able to disrupt PPIs efficiently. The larger molecules and protein-based therapies, including the engineered antibodies, on the other hand, are potent and more selective, but have restricted ability to reach the target tissues and cross the cell membranes. The stapled peptides are large enough to be specific and potent for viral infections to inhibit PPIs and can be modified to enhance their membrane permeability to reach the target cells [60]. The stapled peptides have the potential to act at multiple levels in virus replication cycle. Drug-development-wise, the development of stapled peptides can be expressed and has a short market time.
Blocking CD147 inhibits CoV Replication
The CD147 (basigin), transmembrane glycoprotein belonging to the immunoglobulin superfamily, is important factor in the host defence and regulates the expression of extracellular matrix metalloproteinases (EMMPs). It is expressed in various tissues and cell types, including leukocytes, platelets, epithelial cells and endothelial cells The and fibroblasts. CD147 has been shown to play a prominent role in the induction of pro-inflammatory and pro-thrombotic events in various disease models [61]. The CD147-SP interaction enhances viral invasion for host cells and blocking CD147 on the host cells has shown an inhibitory effect on SARS-CoV-2 p [62]. It has been documented that Meplazumab, a humanized anti-CD147 antibody, can competitively inhibit the binding of S protein and CD147, and prevent the viruses from invading host cells.
Developing/Repurposing Antiviral Drugs
The antiviral agents are being tested in clinical trials.
Remdesivir, a broad-spectrum antiviral and nucleotide prodrug,
inhibits replication of hCoVs in tissue cultures. The anti-retroviral
and other investigational antiviral drugs are being tested for efficacy
against COVID-19. Beta-D-N4-hydroxycytidine (NHC), plitidepsin
and favipiravir are other anti-viral drugs in clinical trials. There
have been efforts for using oligonucleotides against SRS-CoV-2
RNA genome or repurposing currently available various antiviral
medications. The integrative network-based systems pharmacology
methods have been used for rapid identification of repurposable
drugs and drug combinations for the potential treatment of 2019-
nCoV/SARS-CoV-2 [63].
In the cell culture infection models, a cyclophilin inhibitor,
cyclosporin A (CsA), inhibited the replication of CoVs. CsA
binds to cellular cyclophilins to inhibit calcineurin, a calciumcalmodulin-
activated serine/threonine-specific phosphatase,
and CoV replication [64]. The CK2 inhibitors Emodin and Rhein,
anthraquinone compounds and chrysin, a flavonoid compound,
target autophagy via different upstream pathways including the
AKT/mTOR-axis and transcription of autophagy-related proteins.
Whereas, emodin has been shown to block the S protein and ACE2
interaction in a dose-dependent manner. The translation inhibitors
enable the ongoing translation of messenger ribonucleoproteins
(mRNPs) that encode antiviral factors such as interferon-stimulated
genes (ISGs) despite the arrest of bulk translation [65]. Whereas,
the viral transcription inhibitors, like sirolimus, temsirolimus and
everolimus (mTOR inhibitors) block the mTOR, a serine/threonine
protein kinase that regulates protein synthesis, autophagy and
transcription. The toll-like receptor 3 (TLR-3) agonist, rintatolimod
is being tested as a potential treatment for Covid-19 in Japan.
Immuno-modulators and Other Drugs
The interleukin-6 inhibitors, such as tocilizumab (Genentech), may ameliorate severe damage to lung tissue caused by cytokine storm in patients with severe Covid-19. Tradipitant (Vanda Pharmaceuticals), a neurokinin-1 (NK-1) receptor antagonist, may have efficacy for inflammatory lung injury associated with severe SARS-CoV—19 infection. Corticosteroids are not generally recommended for treatment of Covid-19 or any viral pneumonia [66].
Conclusion: Pespectives And Implications
CQ and HCQ in Treating COVID-19
There has been a growing interest in the use of chloroquine (CQ) and hydroxychloroquine (HCQ) as potential treatments in the interim till a specific treatment is available, based on several in vitro and few in vivo studies reporting antiviral activity of CQ and HCQ against SARS-CoV-2 [67]. It has been claimed that HCQ is more potent than CQ against SARS-CoV-2, in vitro [68]. There are some in vivo studies data at present, though limited [69]. A number of potential mechanisms of action of CQ/HCQ against SARS-CoV-2 have been postulated. CQ may reduce glycosylation of ACE2, thereby preventing SARS-CoV-2 from effectively binding to host cells. Furthermore, CQ might block the production of pro-inflammatory cytokines (such as IL-6), thereby blocking the pathway that may lead to acute respiratory distress syndrome (ARDS). CQ is also believed to raise the pH level of the endosome, which may interfere with virus entry and/or exit from host cells.
The prophylactic use of HCQ is not backed-up by research data and trials are underway relating to preexposure and post exposure prophylaxis [70]. Encouraging clinical results were reported from China in February 2020, revealing that the treatment of over 100 patients with chloroquine phosphate in China had resulted in significant improvements of pneumonia and lung imaging, with reductions in the duration of illness [71]. At present, considering all the data, there is insufficient evidence to endorse that CQ/HCQ are safe and effective treatments for Covid-19 [72].
In addition, HCQ has a potential QT-prolonging effect by blocking critical potassium channels in cardiac conduction system, with the possibility of arrythmias and sudden cardiac death. The Covid-19 patients with a baseline QTc value greater than or equal to 500 milliseconds and those that experience an acute QTc reaction with a QTc greater than or equal to 60 milliseconds from baseline after starting treatment with one or more QTc-prolonging drugs are at greatest risk for drug-induced arrhythmias.
Interplay Between Covid-19 and the RAAS
The perspective that ACE inhibitors, like ramipril and ARBs, like losartan may increase ACE2 expression, have raised concerns about their safety in patients with Covid-19. The interaction between the SARS-CoVs and ACE2 has been proposed as a potential factor in their infectivity by some researchers, and there are concerns about the use of these drugs may alter the ACE2 expression and attended by its fallouts [73]. Currently, there are insufficient data available to translate it into clinical practice, though studies are underway. On the other hand, it is held that an abrupt withdrawal of these drugs in high-risk patients, having heart failure, coronary heart disease and hypertension may result in clinical instability and adverse outcomes. Further, despite substantial structural homology between ACE and ACE2, their enzyme active sites are distinct and the ACE inhibitors and ARBs in clinical use do not directly affect the ACE2 activity. Thus, it is advisable that an ACE inhibitor or ARB should be continued in known or likely Covid-19 patients with stable condition [74].
The SARS-CoV-2 utilizes and affects ACE in multiple ways. Apart from its entry through the ACE2, SARS-CoV-2 subsequently downregulate ACE2 expression leading to loss of its protective effects in various organs leading to uncontrolled angiotensin II activity, which may potentiate organ injury in Covid-19. The continued viral infection and replication contribute to reduced membrane ACE2 expression as the ACE2 sites are taken up and damaged by subsequent viral invasions. The reduced ACE2 activity in the lungs results in unopposed angiotensin II accumulation and local RAAS activation, which leads to inflammation and neutrophil infiltration. In the experimental mouse models, exposure to SARS-CoV-1 spike protein induced acute lung injury, which was prevented by RAAS blockade. Similarly, in the Covid-19 patients having elevated levels of plasma angiotensin II, which correlated with viral load and degree of lung injury and the restoration of ACE2 through the administration of recombinant ACE2 appeared to reverse the devastating lung-injury process [73]. SARS-CoV-2 binding to ACE2 creates an acutely exaggerated proinflammatory background and it is advised that clinicians continue treating patients with ACE inhibitors and ARB [75].
Convalescent Plasma and COVID-19 Antibodies
Considering the potential options to treat Covid-19, there have been identified certain proteins in the host immune pathways which can be targeted for blocking viral replication by potential drugs or antibodies [76]. These include antiviral pathways in the innate immune response, such as the stress granule protein, G3BP1, which is an antiviral protein that induces the innate immune antiviral response. The passive antibody transfer from pooled convalescent patient sera is another obvious therapeutic option in severe SRS-CoV-2 infection. The conceptual framework for using it as treatment for Covid-19 has been outlined in two recent review papers [77,78]. To date, two small case series have been published about its use and effectiveness by Chinese researchers [79,80].
Footnotes
1. Affiliation – Senior Consultant and Faculty, Department of
Medicine, Hindu Rao Hospital and NDMC Medical College, New
Delhi, India.
2. Disclosures – None.
3. The Figures 1-3 in this Review Article are subject to
Copyright by Dr Vinod Nikhra.
Competing Interests
The author declare that they have no competing interests.
References
- Masters PS (2006) The Molecular Biology of Coronaviruses. Virus Research 66: 193-292.
- Bergmann CC, Lane TE, Stohlman SA (2006) Coronavirus infection of the central nervous system: host–virus stand-off. Nature Reviews Microbiology 4(2): 121-132.
- Yang P, Wang X (2020) COVID-19: a new challenge for human beings. Cell Mol Immunol.
- Zhou P, Yang XL, Wang XG, Ben Hu , Lei Zhang, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798): 270-273.
- Zhang Y, Zhang C, Zheng W, Eric W Bell, Xiaogen Zhou, et al. (2020) Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. J Proteome Res19 (4): 1351-1360.
- Xiao K, Zhai J, Feng Y, Xiao K, Zhai J, Feng Y, et al. (2020) Isolation and Characterization of 2019-nCoV-like Coronavirus from Malayan Pangolins. Preprints from MedRxiv and bioRxiv.
- Wahba L, Jain N, Fire AZ, Massa J Shoura, Karen L Artiles, et al. (2020) Identification of a pangolin niche for a 2019- nCoV-like coronavirus through an extensive meta-metagenomic search. bioRxiv.
- Lam TT, Shum MH, Zhu H, Yi-Gang Tong, Xue-Bing Ni, et al. (2020) Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature.
- Wang Q, Qiu Y, Li JY, Zhi-Jian Zhou , Ce-Heng Liao, et al. (2020) A Unique Protease Cleavage Site Predicted in the Spike Protein of the Novel Pneumonia Coronavirus (2019-nCoV) Potentially Related to Viral Transmissibility. Virol Sin: 1-3.
- Walls AC, Park YJ, Tortorici MA, Abigail Wall, Andrew T McGuire, et al. (2020) Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell.
- Ye ZW, Yuan S, Yuen KS, Sin-Yee Fung, Chi-Ping Chan, et al. (2020) Zoonotic origins of human coronaviruses. Int J Biol Sci16(10): 1686-1697.
- Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol1282: 1-23.
- Chen Y, Liu Q, Guo D (2020) Emerging coronaviruses: Genome structure, replication, and pathogenesis. J of Med Virology.
- Hulswit RJG, De Haan CAM, Bosch BJ (2016) Coronavirus Spike Protein and Tropism Changes. Adv Virus Res96: 29-57.
- Lu R, Zhao X, Li J, PeihuaNiu , Bo Yang, et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet395(10224): 565-574.
- Song W, Gui M, Wang X, Ye Xiang (2018) Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PlosPathogens.
- Zhou P, Yang XL, Wang XG, Ben Hu, Lei Zhang, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature579(7798): 270-273.
- Schoeman D, Fielding BC (2019) Coronavirus envelope protein: current knowledge. Virol J6:69.
- Hamming I, Timens W, Bulthuis MLC, A T Lely, G J Navis, et al. (2004) Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol203(2): 631-637.
- Xu H, Zhong L, Deng J, Jiakuan Peng, Hongxia Dan, et al. (2020) High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci12:8.
- Xie CJ, Wang X, Zhang F, Furong Zhang, Yanrong Liu (2006) Age- and gender-related difference of ACE2 expression in rat lung. Life Sci78(19): 2166-2171.
- Chen J, Jiang Q, Xia X, Kangping Liu, ZhengqingYu ,et al. (2020) Individual Variation of the SARS-CoV2 Receptor ACE2 Gene Expression and Regulation. Preprints.
- Lekto M, Marzi A, Munster V (2020) Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature microbiology5: 562-569.
- Chan JF, Yuan S, Kok KH, Kelvin Kai-Wang To, Hin Chu, et al. (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet395: 514-523.
- Jia HP, Look DC, Shi L, Lecia Pewe, Jason Netland, et al. (2005) ACE2 Receptor Expression and Severe Acute Respiratory Syndrome Coronavirus Infection Depend on Differentiation of Human Airway Epithelia. J Virol79(23): 14614-14621.
- Zhang H, Penninger JM, Li Y, Nanshan Zhong, Arthur S Slutsky, et al.(2020) Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Medicine 46: 586-590.
- (2004) Cumulative Number of Reported Probable Cases of Severe Acute Respiratory Syndrome (SARS). WHO (2004) SARS.
- World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV).
- Munster VJ, Koopmans M, Van Doremalen N, Debby van Riel, Emmie de Wit (2020) A Novel Coronavirus Emerging in China - Key Questions for Impact Assessment. N. Engl. J. Med 382: 692-694.
- Loo YM, Gale M (2011) Immune signaling by RIG-I-like receptors. Immunity34(5): 680-692.
- Lazarte JMS, Thompson KD, Jung TS (2019) Pattern Recognition by Melanoma Differentiation-Associated Gene 5 (Mda5) in Teleost Fish: A Review. Front. Immunol.
- Bruns AM, Horvath CM (2015) LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling. Cytokine74: 198-206.
- De Wilde AH, Snijder EJ, Kikkert M, Martijn J van Hemert(2018)Host Factors in Coronavirus Replication. Curr Top Microbiol Immunol419: 1-42.
- Yang D, Leibowitz JL(2015) The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res206: 120-133.
- Bhardwaj K, Liu P, Leibowitz JL, C Cheng Kao (2012) The Coronavirus Endoribonuclease Nsp15 Interacts with Retinoblastoma Tumor Suppressor Protein. J Virol86(8): 4294–4304.
- Ma-Lauer Y, Carbajo-Lozoya J, Hein MY, Marcel A Müller, Wen Deng, et al. (2016) p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. PNAS Plus Microbiology, Proc Natl Acad Sci U S A. 113(35): E5192-E5201.
- Chang JW, Zhou YQ Qamar, Ling-Ling Chen, Yu-Duan Ding (2016) Prediction of Protein–Protein Interactions by Evidence Combining Methods. Int. J. Mol. Sci17(11): 1946.
- Brito AF, Pinney JW (2017) Protein–Protein Interactions in Virus-Host Systems. Front. Microbiol.
- Franzosa E, Xia Y (2011) Structural principles within the human-virus protein-protein interaction network. Proceedings of the National Academy of Sciences108(26):10538-10543.
- Selinger C, Tisoncik-Go J, Menachery VD, Sudhakar Agnihothram, G Lynn Law, et al. (2014) Cytokine systems approach demonstrates differences in innate and pro-inflammatory host responses between genetically distinct MERS-CoV isolates. BMC Genom 15:1161.
- Emmott E, Munday D, Bickerton E, Mark A Rodgers, Adrian Whitehouse, et al.(2013) The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J Virol 87: 9486-9500.
- Bi J, Song S, Fang L, Dang Wang, Huiyuan Jing, et al. (2014) Porcine Reproductive and Respiratory Syndrome Virus Induces IL-1β Production Depending on TLR4/MyD88 Pathway and NLRP3 Inflammasome in Primary Porcine Alveolar Macrophages. Mediators Inflamm pp.403515.
- Shi C, Nabar NR, Huang N, John H Kehrl (2019) SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov5: 101.
- Liu J, Liu Y, Xiang P, Lin Pu, HaofengXiong, et al (2020) Neutrophil-to-Lymphocyte Ratio Predicts Severe Illness Patients with 2019 Novel Coronavirus in the Early Stage. 2020, Preprint from medRxiv and bioRxiv.
- Yang Y, Peng F, Wang R, Kai Guan, Taijiao Jiang, et al.(2020) The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmunity109:102434.
- Chen IY, Moriyama M, Chang MF, Takeshi Ichinohe (2019)Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front Microbiol 10:50.
- Huang C, Wang Y, Li X, Lili Ren, Jianping Zhao, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet395(10223):497-506.
- Mao L, Wang M, Chen S, Quanwei He, Jiang Chang, et al. (2020) Neurological Manifestations of Hospitalized Patients with COVID-19 in Wuhan, China: a retrospective case series study. 2020, Preprint from medRxiv and bioRxiv.
- Mesev EV, LeDesma RA, Ploss A (2019) Decoding type I and III interferon signalling during viral infection. Nat Microbiol4(6): 914-924.
- Chen WH, Strych U, Hotez, Bottazzi ME (2020) The SARS-CoV-2 Vaccine Pipeline: An Overview. Curr Trop Med Rep p.1-4.
- Coleman CM, Liu YV, Haiyan M, Taylor JK, Massare M, et al. (2014) Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine 32(26): 3169-3174.
- (2020) Clover Biopharmaceuticals. Clover initiates development of recombinant subunit-trimer vaccine for Wuhan coronavirus (2019-nCoV).
- Chen WH, Chag SM, Poongavanam MV, Mohan V Poongavanam, Amadeo B Biter, et al. (2017) Optimization of the production process and characterization of the yeast-expressed SARS-CoV recombinant receptor-binding domain (RBD219-N1), a SARS vaccine candidate. J Pharm Sci106(8): 1961-1970.
- Hoffmann M, Kleine-Weber H, Schroeder S, Nadine Krüger, Tanja Herrler, et al. (2020) SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell.
- Zhou Y, Vedantham P, Lu K, Juliet Agudelo, Ricardo Carrion, et al.(2015) Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Research116: 76-84.
- Liu W, Fontanet A, Zhang PH, Lin Zhan, Zhong-Tao Xin, et al. (2006) Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J. Infect. Dis193: 792-795.
- Kuba K, Imai Y, Rao S, Hong Gao, Feng Guo, et al.(2005) A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat Med 11(8): 875-879.
- Imai Y, Kuba K, Penninger JM (2008) The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice. Exp Physiol 93(5):543-548.
- Battle D, Wysocki J, Satchell K (2020) Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci (Lond) 134 (5): 543–545.
- https://blog.dana-farber.org/insight/2020/04/pioneering-a-staple-approach-for-treating-the-coronavirus-covid-19/
- Wang K, Chen W, Zhou YS, Jian-Qi Lian, Zheng Zhang, et al. (2020) SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. March 2020, Preprint from medRxiv and bioRxiv.
- Heinzmann D, Noethel M, Von Ungern-Sternberg S, IoannisMitroulis, Meinrad Gawaz, et al.(2020) CD147 is a Novel Interaction Partner of Integrin αMβ2 Mediating Leukocyte and Platelet Adhesion. Biomolecules10: 541.
- Zhou Y, Hou Y, Shen J, Yin Huang, William Martin, et al. (2020) Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov.
- Tanaka Y, Sato Y, Sasaki T (2013) Suppression of Coronavirus Replication by Cyclophilin Inhibitors. Viruses5(5): 1250-1260.
- Zhang L, Lin D, Kusov Y, Yong Nian, Qingjun Ma, et al. (2020) α-Ketoamides as Broad-Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, Synthesis, and Activity Assessment. JMedChem.
- (2020) Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected. World Health Organization.
- Yao X, Ye F, Zhang M, Cheng Cui, Baoying Huang, et al.(2020) In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clinical Infectious Diseasespii: ciaa237.
- Liu J, Cao R, Xu M, Xi Wang,Huanyu Zhang, et al. (2020) Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discovery6(1): 1-4.
- Gautret P, Lagier JC, Parola P, Van Thuan Hoang, Line Meddeb, et al.(2020) Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents.
- (2020) WebMD Health News. Karen Weintraub. Chloroquine, Zinc Trials Underway for COVID-19 Prophylaxis.
- Gao J, Tian Z, Yang X (2020) Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends14(1):72-73.
- Gbinigie K, Frie K (2020) Should chloroquine and hydroxychloroquine be used to treat COVID-19? A rapid review. BJGP.
- Vaduganathan M, OrlyVardeny O, Thomas Michel T, John JV McMurray, Marc A Pfeffer,et al. (2020) Renin–Angiotensin–Aldosterone System Inhibitors in Patients with Covid-19. NEJM.
- AlGhatrif M, Cingolani O, Lakatta EG (2020) The Dilemma of Coronavirus Disease 2019, Aging, and Cardiovascular Disease: Insights from Cardiovascular Aging Science. JAMA Cardiol.
- Pal R, Bhansali A (2020) COVID-19, diabetes mellitus and ACE2: The conundrum. Diabetes Res Clin Pract162: 108132.
- Gordon DE, Jang GM, Bouhaddou M,Jiewei Xu,Kirsten Obernier, et al. (2020) A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. Preprint from medRxiv and bioRxiv.
- Casadevall A, Pirofski LA (2020) The convalescent sera option for containing COVID-19. J Clin Invest130 (4):1545-1548.
- Bloch EM, Shoham S, Casedevall A, Bruce S Sachais , Beth Shaz, et al.(2020) Deployment of convalescent plasma for the prevention and treatment of COVID-19. J Clin Invest.
- Shen C, Wang Z, Zhao F, Yang Yang, Jinxiu Li, et al.(2020) Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA.
- Duan K, Liu B, Li C,Huajun Zhang, Ting Yu, et al. (2020) Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci USA.

 Review Article
Review Article