Abstract
Integrated cancer patients profiling using combinations of PDC-PDx-Omics technologies are receiving growing scientific interest as a strong approach for clinical decisional workflows leading to personalized treatment. Nonetheless enforcing such workflows requires extensive collection and extended availability of the patients bioasset (specimen plus personal health and genomic data). However, biobanking and data collection from a large number of individuals bear both cost sustainability and ethical issues limiting the adoption of deep profiling workflows. These hindrances are linked to the cost and limited sharing of the fragmented patient data/specimen throughout the healthcare providers and medical research systems along with the challenges in attracting large numbers of informed and consenting patients in biobanking programs, as they are offered today. Starting from the game-changing potential in the inclusion of PDC-PDx-Omics based and A.I.-driven decisional workflows in routine precision oncology, the present article considers the practical obstacles for the use of this approach in current practices. Furthermore, we analyze the present trends in the type of operational environments used to deliver PDC-PDx-Omics-based PGM services and provide new perspectives on the potential adoption of a non-profit, expert-patients-forpatients- driven, A.I.-backed, biorepository-CRO operational model towards attaining financial sustainability, wider service access and ethically compliant deep patients profiling towards effective cures.
Abbreviations: PDC: Patient-Derived Cells, PDX: Patient-Derived Xenograft, CAF: Cancerassociated fibroblasts, Omics, Genomic/Transcriptomic/Proteomic technologies; NOD/ SCID, Non Obese Diabetic/Severe Combined Immune-Deprived; PGM: Personalized Genomic Medicine, DTC: Direct-To-Client, CRO: Contracted Research Organization, FFPE: Formalin-Fixed Paraffin-Embedded, AI: Artificial Intelligence, ELSI: Ethical-Legal-and Social Implication, IMI: Innovative Medicines Initiative.
Introduction
The Role of PDC-PDx-Omics Studies in Precision Oncology: towards Cancer Cure in each Patient
The power of large cohorts’ cancer genomic studies in setting the baseline between normal and cancer-driving mutational events has recently been further demonstrated by the results of the pan-cancer project [1]. Importantly the study, which included more than 2600 whole cancer genomes has shown that although several cancer-driving mutations can occur the non-coding DNA regions, the vast majority of cancer-driving mutations occurs on the protein-coding regions [2]. On the other hand, the realization that potentially more than 57% of cancer-driving mutations l are druggable in patients including those mutations acquired after starting a cancer treatment regimen [3], along with the understanding that PDCs can be consistently obtained upon PDx expansion [4], strengthens the rational for the adoption of these integrated workflows in the personalized genomic-driven clinical oncology setting. A key role in the establishment of fully personalized curative solutions, therefore, relies upon the advances of PDC-based technologies for biologic, phenotypic, and proteo-genomic profiling.
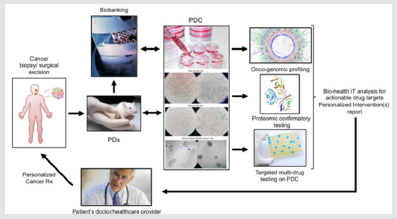
This explains the intense research investigation activity witnessed in the recent years meant to optimize the collection, expansion and organotypic culture of patients derived cancer cells in order to successfully apply the above mentioned ex-vivo testing to the individual patient [5] as also summarized in Figure 1. According to this research, innovative knowledge is available towards optimization of PDC establishment efficiency from (a) circulating tumor cells [6,7], (b) tumor biopsies [8], (c) tumor-associated ascitic fluid [9] as well as to 3D culture establishment both in spheroid (single cell line) and organoid (PDC+CAF or other stromal component) format [10]. The Patient Derived Xenograft (PDx) testing can be considered as medical application of the established mouse xenograft tumorigenic assay involving the implantation and study of tridimensional growing human cancer cells in in the sub- cutaneous tissue of genetically immune-deprived mice [11]. Cancer Xenografts have been made possible following the identification of a severe immunocompromised deficiency syndrome in a mouse [12] conveyed to the development of the NOD/SCID mouse [13], a genetically engineered mice model that minimizes or avoids Host versus Graft immunologic rejection upon surgical implantation of cultured human cancer cells [14-17] or bioptic tumor tissue [18].
This tumorigenic in vivo assay has helped advancing our biologic knowledge of cancer features undetectable in 2D cultures such as tumor vascularization [19-21] and metastasis [22,23] as well as providing a powerful assay to study anti-tumorigenic actions of drugs on tridimentionally-growing tumoral cells typical of solid cancers. However even NOD/SCID mice would maintain a quote of immunological resistance to human cells, namely the natural killer- dependent activity. Hence, investigators further engineered a mouse strain harboring a mutation ablating such mechanism conveying it to what is ultimately known as the NSG (NOD SCID Gamma) mouse, which constitutes the gold standard for conducting PDx studies [24,25], including a humanized strain suitable for immuno- oncology studies [26]. At the technical level, the combination of the PDC and the PDx technology overcomes the limitation in the number of starting bioptic cancer tissue allowing a variety of tests to be included and standardized in the proposed workflow. In fact, a single pathological cell or a small number of cells obtained from the patient during a biopsy (tissue surgery, a fine needle aspiration, brushing, etc) can be used to generate a patient-derived cell line that can be first expanded in vitro, eventually grown in 3D culture (to generate a spheroid or organoid) and consequently re-implanted in the PDx animal for expansion.
This step has been demonstrated to be minimally affected by possible acquired mutations and not in ways that would affect the major cancer-driving and phenotypic abnormalities of the primary PDC [24]. Following this last step, this tissue can be explanted to generate a secondary tissue patient-specific line and patient cells can be tested in High-throughput format towards genomic, proteo- transcriptomic and multi-drug personalized screening (Figure 1). This approach has been shown to be able to have a therapeutic impact on the cancer patient throughout the course of his disease [27]. Summarizing the general scientific consensus for the PDCPDx- driven workflow, in 2017, a group of experts at the Memorial Sloan Kettering Cancer Center, has suggested that the combination of tissue biopsy, liquid biopsy, patient-derived organoids establishments and patient-derived xenograft could, indeed, be considered as an integrated routine approach towards target confirmation, target engagement, target validation and acquired resistance identification in each subject [28].
These highlighted aspects should be considered as part of a real-time investigative process meant to benefit the individual patient but also to provide valuable integrated data and rational for the treatment of other patients sharing comparable molecular and genomic alterations. Given the unlimited value linked to the biorepository and personal big genomic data ownership by genomic service providers in the international landscape, specific solutions and considerations on the most suitable and anti-trust compatible operational model for the implementation of these large biologic and big-data collectors is highly invoked.
Ethical and Financial Model Restrictions to Personalized Big (bio)Data and Bio- Specimen Collection Generated via the PDC-PDx and Multi-Omics Testing: The Patient is NOT the Enemy
There is universal consensus among experts and scientists on the advantage and/or need to share individual patient health and biogenomic data towards implementing the power of genomic- driven medicine. However, the parallel hot topic related to individual patients right to privacy and/or the validity of the individual informed consent for the unrestricted use of personal (bio genomic) data has been addressed internationally in ways meant to simply change the individual mind as far as the advantage/need to educate ourselves to give out our personal consent without enforceable guarantees on the personal benefit and actual negotiating options towards the best possible personal outcome [29,30]. In other cases, there has been a rationalized attempt in depriving patients of the power of consent for genomic medical testing [31] in spite a part of the international inter-disciplinary experts favors the patients direct involvement in the fate of their own health and genomic data [32-35]. Not less controversial is the involvement of private parties which have already shown predatory collection practices of patient’s personal health information in a partially and/or bias informed fashion [36], especially by the dot com industry [37-39]. Similar concerns include the lack of personal guarantees and control over the use of personal bio-asset by any private corporate Direct- To-Consumer (DTC) genomic provider [40-42].
A.I. & PDC-PDx-Omics: Natural Partners
The need for rapid standardization in evidence-based medicine at the exponentially growing pace of scientific progress in the bioinformatic, biomolecular and biomedical post-genomic fields, has been a major driver for the introduction and use of deep-learning neural networks [43]. The A.I.-based approaches play a major and exponentially growing role in the analysis of big-data derived from a single patient and compare them using expert-curated algorithms to large cohorts of clinical data from other patients. In particular, they are essential in the collection, functional organization, validation and smart analysis of big data generated through the Omics and high-input/throughput medical and health data digital devices providing either quantitative (bit) or imaging (pixel) data and apply translational scientist/clinician-curated algorithms in order to provide both visual summaries and infographics in real time to generate corrective medical actions and/or specific health queries with immediate and, at times, life-saving medical interventions. Among these, at present, are early detection of heart/brain acute conditions, early diagnosis and treatment monitoring of a variety of medical conditions and the detection of genetic predisposing and or adverse complex genetic conditions [44-51].
In this regard, the use of A.I.-driven technology to any PDC-PDx- Omics integrated process offers potential unique opportunities starting from the standardization of PDC-PDx-Omics-based workflow in each patient since the QA/QC and smart data enrichment/ flagging process requested for obtaining efficient and cost-effective profiling and bio-asset (data and specimen) repository management are a major hindrance to both the sustainability and potential growth of current biobanking services towards wider access to personalized PDC-PDx- based profiling as well as representing an additional limit to the adoption of such practice at the international level. However, in spite of the above mentioned pros, the one single most relevant advantage, where A.I. could prove itself bringing neutrality and speed up the adoption of the PDC-PDx-Omics workflow in current oncology practices, is the intrinsic capability to anonymize, encrypt, decode and protect individual personal and genomic identity. This is even more relevant in the digitally integrated world we fully dive in as we come to the attention of a healthcare provider because of an oncological problem. For this reason, we believe that A.I. constitutes an intrinsic tool for the proposed no-profit biobank-CRO operational model.
Is the PDC-PDx-Omics Testing a Sustainable Option for Routine Precision Oncology?
The answer to this question is “not as it is”. In fact, at present the potential cost for the use of PDC-PDx-Omics workflow in routine oncology would increase the overall individual diagnostic costs to the range of several thousands of US dollars per single patient. It is worth noticing that, although the identity and number of cancer driver genes has been identified [52-55], there is not yet a general consensus on the necessary or minimal standard use of genomic,- transcriptomic and proteomic testing to be pursued on a single patient, and to the dept of the Omics profiling to be brought to the clinical level. On the practical standpoint, the traditional pathological techniques based on tissue fixation constitute a hindrance to the collection of the patient’s living cancer cells for PDC-PDx testing. In fact, most institutions current PGM workflow relies on FFPE-based Omics tests which cannot guarantee living primary tumor cells expansion and specific functional characterization such as the multidrug test on PDC on a regular basis (Figure 1) which has already demonstrated its therapeutic advantages [27].
Under these circumstances, the cost for implementation of the current clinical practices as well as to the introduction of alternative operational models to be used in precision oncology offers a topic for open discussion. In such international landscape, based on the initiatives of each institution backed by the personalized medicine Dx and Rx stakeholders, and institution-owned biobanks, the patients option to obtain best possible genomic and highly discriminative genetic-embedded biologic data analysis is undermined in exchange of a simple consent signature locking the individual bio-asset to that institution almost irreversibly (even worse when the patient or apparently healthy individual submits his/her bioasset to any corporate DTC provider with practically no guarantee of safety or control over the use and profiling of his/her own biologic data and material). Furthermore, once made the (currently only available) choice of giving out completely ones’ own biologic self (the personal genome), to a corporate entity, this makes the individual’s rights highly difficult if not impossible to protect and enforce due to the disparity of the entities at stake.
The Expert-Patient Potential Contribution to Unleash the PDC-PDx-Omics Model
A potential effective solution to the hindrance determined by the corporate- centered dynamics and operational models mentioned above stands on the simple consideration that medical and multidisciplinary scientists with unique experience and knowledge working in non-lucrative research laboratories could be presently, have been or will be affected by the same diseases that corporate- driven/Institution-centered entities do treat. In spite of this apparently obvious consideration pointing at the fact that a number (although limited) of cancer patients are actually involved in the same cancer-fighting jobs and activities throughout the entire spectrum of current stakeholders, the potential role of such sub-group of individuals towards promoting/educating the general population to the safe use of biobanking towards health advantages is currently underestimated. Indeed, we believe their role (as managing part of patients for patients-driven non lucrative organizations) could represent a solution in order to abate the above-mentioned restrictions.
Basically, Institution-based operational models put the profit and growth of the individual institution above the specific interests of the individual patient (since the patient is not a rightful stakeholder of his bioasset but a mere donor/voluntary client instrumental to the growth and gain of the service providing Institution. A key limit with Institution-centered/corporate-driven PGM model is the intrinsic high cost of the testing (as can be stated of the PDCPDx- Multi-Omics-driven Oncology) which is affordable to a very limited sub-group of patients affected by the disease. Due to the very large number of the individuals projected to develop cancer at the global level in the next decade [56-58], when considering the restricted access to Omics-driven personalized treatments possible under the current cancer PGM healthcare operational model, the actual advances in PDC-PDx-Omics will poorly if not negligibly affect the outcome of the disease. The profound limitations of the current oncology Dx-Rx workflow had been already highlighted by the WHO in 2005 [59] and by relevant Oncological organizations (such as ASCO, ACS, UICC) since 2010 [60].
This led to the Precision Medicine initiative and the increase in expenditures to further develop and/or perfect genomic sequencing and extend its use in the clinical setting. Nonetheless, an operational biobanking model alternative to the corporate-based genomic service collector model has remained as the sole vehicle of personalized oncology. The problem having a corporate- for profit genomic-service provider manage patients’ biobanks, while using its own brand and resources to compete with other institutions for limited bio-assets, results in the intrinsic exclusive use of the patient- derived specimen for intra-mural research. This “silo-effect” provides the institution’s own translational research with unfair advantages let alone depriving the patient for alternative options beneficial to his own health and it further contributes to the current fragmentation of the patient bio-records and bio-specimen throughout the healthcare system avoiding any possible drop in the cost of personalized genomics. The involvement of expert patients- driven no-profit biobanks as a possible solution to preventing/ eliminate the ethical hidden risk and mass (patients’) rights infringements deriving from the use of PDC-PDx-Omics services under corporate-managed and Institution-centered Biobanks could indeed lead to transparent CRO practices that could fulfil the need and scopes of both PGM stakeholders and individual patients without any change in current patient privacy international regulations.
Furthermore, the involvement of Patients-driven Biobanks can provide an extremely valuable source of patient’s population able to minimize the unwanted bias provided by early genomic cohorts studies. In the US, the NIH is promoting research towards the ethical implications of the use of genomics in human medical and non-medical applications through ELSI [61,62]. However, to date, the results of the research promoted through such channel towards the establishment of sustainable operational models for genomic medicine in relationship to the involvement and/or role of the patient in the PGM operational models are vague.
Advantages of Expert Patients/A.I.-driven No Profit Biobanks in Supporting Institution-Centered Precision Cancer Medicine Services
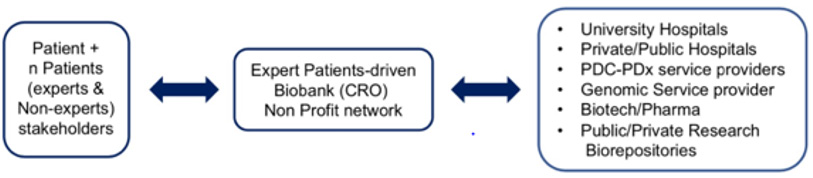
As summarized in (Figure 2), a possible game-shifting opportunity to implement the pure institution based operational model to a more patient-active/compliant financially and ethically sustainable model to deliver PDC-PDx-multi-Omic services can come in our view from the inclusion of expert patients-for-patients biobanks/ CROs operating under no-profit status for the reasoning previous ly discussed. A typical patient bio-asset (specimen + data) benefiting from A.I.- backed virtual integration and analysis of present as well as retroactive data obtained from each individual patient/ stakeholder includes the patient clinical and biomarkers testing history, a cancer/chronic diseases genetic family tree and genetic counseling reports, genomic testing reports (ranging from targeted/ single gene, transcriptomic to WGS data) and molecular imaging files (body/tissue level). In summary, the key principle enabling such model is that each basic and translational scientist as well as top professionals in multidisciplinary areas are actual or potential patients. As such, such (current or foreseeable) expert patients are stakeholders of their own bio-asset and have the proper incentive to represent non-technical patients’ rights, provide expert-curated services to improve and update the rapidly evolving clinical and bio-genomic knowledge- based assets, as well as guarantee the anonymization and long term preservation of the individual’s community biodata.
Figure 2: Proposed sustainable PDC-PDx-Omics workflow in Precision Oncology via adoption of an Expert Patient-driven no-profit biobank-CRO network.
Furthermore, by acting under the proposed entity model they represent an ideal interface with both academic and corporate dynamics by making each patient bio-asset (properly anonymized and QA/QC-validated) available to the patient-selected PDC-PDx-Omics healthcare providers therefore solving the current problem of the fragmentation of the patient bio-asset among several competing institutions which currently represents the major cost-barrier to establish wider personalized PDC-PDx-Omics-driven programs. Although there are only a few examples of established no profit research/service organizations operating in the field of PDC and/ or PDx and/or Omics services, none have adopted (so far) an Expert- Patient-For-Patient framework. We believe that the adoption of such operational model has unexplored and highly underscored positive implications towards the spreading of this life-saving workflow.s
Conclusion and Perspectives
Based upon the level of scientific views and validation in the personalized oncology field, PDC-PDx-Omics services are set to keep growing exponentially in the international landscape. Such growth at present is currently linked to an Institution-centered, corporate-driven biobanking model having as parallel effect the undisclosed transfer of ownership of the individual patient bio-asset to multiple institutions. The stainability of this model linked to the patient’s virtual and physical data/specimen fragmentation throughout the various competing PDC-PDx-Omics-driven stakeholders is financially far from sustainable. In fact, the cost of technology and infrastructure involved for each patient profiling, data management and biobanking clearly limits the access of such deep-personalized services and life-saving interventions to a wider number of cancer-affected individuals (in spite of the expected growth in cancer cases in the upcoming decades). The introduction of an Expert-Patient-For-Patients (EPFP) framework as proposed operational model for no profit biobanks/CRO meant as buffer entities for the optimal management/anonymization of personal data, their validation and patients’ specimen standardization/ storage alternative to the corporate for- profit institution-centered biobanking model, has a number of advantages.
First, it can serve in parallel both the individual patient and his due right to health data anonymization in exchange for his/ her shared bio-asset as well as fulfilling the pharma/ corporate/ academic stakeholders goals as users of highly reliable sources of validated data/specimen. At last, but not less important, such model can lead to a substantial abatement of current and foreseeable costs for access to PDC-PDx-Omics services at the final user. Both these effects would guarantee the increase in the adoption of PDCPDx- Omics workflows supporting the spreading of true personalized and curative treatment of cancer and other diseases.
References
- Reyna MA, Haan D, Paczkowska M, Verbeke LPC, Vazquez M, et al (2020) Pathway and network analysis of more than 2500 whole cancer genomes. Nature communications 11: 729.
- Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, et al. (2020) Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 578: 102-111.
- Scalia P, Williams SJ, Russo A, Giordano A (2017) Actionable Molecular Targets in Cancer liquid biopsy In Liquid Biopsy in Cancer Patients: the hand lens to investigate tumor evolution, Russo A, Giordano A, Rolfo C, (Eds.)., Springer International Publishing.
- Dangles Marie V, Pocard M, Richon S, Weiswald LB, Assayag, et al. (2007) Establishment of human colon cancer cell lines from fresh tumors versus xenografts: Comparison of success rate and cell line features. Cancer research 67(1): 398-407.
- Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, et al. (2012) Patient-derived tumour xenografts as models for oncology drug development. Nature reviews Clinical oncology 9(6): 338-350.
- Yu M, Bardia A, Aceto N, Bersani F, Madden MW, et al. (2014) Cancer therapy Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345(6193): 216-220.
- Morrow CJ, Trapani F, Metcalf RL, Bertolini G, Hodgkinson CL, et al. (2016) Tumourigenic non-small-cell lung cancer mesenchymal circulating tumour cells: A clinical case study. Annals of oncology: official journal of the European Society for Medical Oncology 27(6): 1155-1160.
- Kodack DP, Farago AF, Dastur A, Held MA, Dardaei L, et al. (2017) Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care. Cell reports 21(11): 3298-3309.
- Park SY, Kim C, Jang NK, Park J, K Yi, et al. (2015) Patient-derived cell models as preclinical tools for genome- directed targeted therapy. Oncotarget 6(28): 25619-25630.
- Truong DD, Kratz A, Park JG, Barrientos ES, Saini T, et al. (2019) A Human Organotypic Microfluidic Tumor Model Permits Investigation of the Interplay between Patient-Derived Fibroblasts and Breast Cancer Cells. Cancer research 79(12): 3139-3151.
- Shultz LD, Goodwin N, Ishikawa F, Hosur V, Lyons BL, et al. (2014) Human cancer growth and therapy in immunodeficient mouse models. Cold Spring Harbor protocols (7): 694-708.
- Custer RP, Bosma GC, Bosma MJ (1985) Severe combined immunodeficiency (SCID) in the mouse Pathology, reconstitution, neoplasms. The American journal of pathology 120: 464-477.
- Prochazka M, Gaskins HR, Shultz LD, Leiter EH (1992) The nonobese diabetic scid mouse: Model for spontaneous thymomagenesis associated with immunodeficiency. Proceedings of the National Academy of Sciences of the United States of America 89(8): 3290-3294.
- Reddy S, Piccione D, Takita H, Bankert RB (1987) Human lung tumor growth established in the lung and subcutaneous tissue of mice with severe combined immune deficiency. Cancer research 47: 2456-2460.
- Taylor CW, Grogan TM, Lopez MH, Leong SP, Odeleye A, et al. (1992) Growth and dissemination of human malignant melanoma cells in mice with severe combined immune deficiency. Laboratory investigation a journal of technical methods and pathology 67(1): 130-137.
- Itoh T, Shiota M, Takanashi M, Hojo I, Satoh H, et al. (1993) Engraftment of human non-Hodgkin lymphomas in mice with severe combined. immunodeficiency Cancer 72(9): 2686-2694,
- Tong AW, Huang YW, Zhang BQ, Netto G, Vitetta ES, et al. (1993) Heterotransplantation of human multiple myeloma cell lines in severe combined immunodeficiency (SCID) mice. Anticancer research 13(3): 593-597.
- Kubota T, Yamaguchi H, Watanabe M, Yamamoto T, Takahara T, et al. (1993) Growth of human tumor xenografts in nude mice and mice with severe combined immunodeficiency (SCID). Surgery today 23: 375-377.
- Gray DR, Huss WJ, Yau JM, Durham LE, Werdin ES, et al. (2004) Short-term human prostate primary xenografts: An in vivo model of human prostate cancer vasculature and angiogenesis. Cancer research 64(5): 1712-1721.
- Smith V, Wirth GJ, Fiebig HH, Burger AM (2008) Tissue microarrays of human tumor xenografts: characterization of proteins involved in migration and angiogenesis for applications in the development of targeted anticancer agents. Cancer genomics & proteomics 5(5): 263-273.
- Zhou Z, Christofidou-Solomidou M, Garlanda C, DeLisser HM (1999) Antibody against murine PECAM-1 inhibits tumor angiogenesis in mice. Angiogenesis 3: 181-188.
- Naramura M, Gillies SD, Mendelsohn J, Reisfeld RA, Mueller BM (1993) Therapeutic potential of chimeric and murine anti-(epidermal growth factor receptor) antibodies in a metastasis model for human melanoma. Cancer immunology, immunotherapy: CII 37: 343-349.
- Goldstein SD, Hayashi M, Albert CM, Jackson KW, Loeb DM (2015) An orthotopic xenograft model with survival hindlimb amputation allows investigation of the effect of tumor microenvironment on sarcoma metastasis. Clinical & experimental metastasis 32(7): 703-715.
- Izumchenko E Paz, Ciznadija K, Sloma D, Katz I (2017) Patient-derived xenografts effectively capture responses to oncology therapy in a heterogeneous cohort of patients with solid tumors. Annals of oncology: official journal of the European Society for Medical Oncology 28(10): 2595-2605.
- Zhang F, Wang W, Long Y, Liu H, Cheng, et al. (2018) Characterization of drug responses of mini patient-derived xenografts in mice for predicting cancer patient clinical therapeutic response. Cancer Commun (Lond) 38(1): 60.
- Yao LC, Aryee KE, Cheng M, Kaur P, Keck JG, et al. (2019) Creation of PDX-Bearing Humanized Mice to Study Immuno-oncology. Methods Mol Biol 1953: 241-252.
- Chia S, Low JL, Zhang X, Kwang XL, Chong FT, et al. (2017) Phenotype-driven precision oncology as a guide for clinical decisions one patient at a time. Nature communication 8: 435.
- Hyman DM, Taylor BS, Baselga J (2017) Implementing Genome-Driven Oncology. Cell 168(4): 584-599.
- Geneske J CDC Director on What Public Health Teaches Us about Resilience Foundation.
- (2013) The Rockefeller Foundation Innovation Forum, Building Urban Resilience - Dr Tom Frieden You tube.
- Johnson SB, Slade I, Giubilini A, Graham M (2019) Rethinking the ethical principles of genomic medicine services. European journal of human genetics EJHG 28: 147-154.
- Gu W, Yildirimman R, Van der Stuyft E, Verbeeck D, Herzinger S, et al. (2019) Data and knowledge management in translational research: Implementation of the eTRIKS platform for the IMI Onco Track consortium. BMC bioinformatics 20(1): 164.
- Teare HJA, De Masi F, Banasik K, Barnett A, Herrgard S, et al. (2018) The governance structure for data access in the DIRECT consortium: An innovative medicine initiative (IMI) project. Life science society and policy 14(1): 20.
- Klungel OH, Kurz, X, De Groot MC, Schlienger RG, Tcherny Lessenot S, et al. (2016) Multi-centre, multi- database studies with common protocols: lessons learnt from the IMI PROTECT project. Pharmacoepidemiology and drug safety 25 (Suppl 1): 156-165.
- Reynolds RF, Kurz X, De Groot MC, Schlienger RG, Grimaldi Bensouda L, et al. (2016) The IMI PROTECT project: Purpose, organizational structure, and procedures. Pharmacoepidemiology and drug safety 25(Suppl 1): 5-10.
- Davis J (2019) The 10 Biggest Healthcare Data Breaches of 2019, So Far. xtelligent Healthcare Media: Health IT Security.
- (2018) BUSINESS WIRE. Amazon, Berkshire Hathaway and JPMorgan Chase & Company to partner on US employee healthcare. Business Wire.
- (2009) Facebook. Facebook Launches New Preventative Health Tool.
- (2019) HIPPA Journal. Google Confirms it has Legitimate Access to Millions of Ascension Patients’ Health Records.
- Kilbride MK, Domchek SM, Bradbury AR (2018) Ethical Implications of Direct-to- Consumer Hereditary Cancer Tests. JAMA oncology 4: 1327-1328.
- Stoekle HC, Mamzer Bruneel MF, Vogt G, Herve C (2016) 23 and Me: A new two- sided data-banking market model. BMC medical ethics 17: 19.
- Stoekle HC, Mamzer Bruneel MF, Frouart CH, Le Tourneau C, Laurent Puig P, et al. (2018) Molecular Tumor Boards: Ethical Issues in the New Era of Data Medicine. Science and engineering ethics 24(1): 307-322.
- Topol EJ (2019) High-performance medicine: the convergence of human and artificial intelligence. Nature medicine 25: 44-56.
- Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, et al. (2011) Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Science translational medicine 3(108): 108-113.
- Giger ML (2018) Machine Learning in Medical Imaging. Journal of the American College of Radiology: JACR 15: 512-520.
- Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, et al. (2016) Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. Jama 316: 2402-2410.
- Gao M, Igata H, Takeuchi A, Sato K, Ikegaya Y (2017) Machine learning-based prediction of adverse drug effects: An example of seizure-inducing compounds. Journal of pharmacological sciences 133: 70-78.
- Mueller A, Candrian G, Kropotov JD, Ponomarev VA, Baschera GM (2010) Classification of ADHD patients based on independent ERP components using a machine learning system. Nonlinear Biomed Phys 4 (Suppl 1): S1.
- Ahmad T, Lund LH, Rao P, Ghosh R, Warier P, et al. (2018) Machine Learning Methods Improve Prognostication, Identify Clinically Distinct Phenotypes, and Detect Heterogeneity in Response to Therapy in a Large Cohort of Heart Failure Patients. Journal of the American Heart Association 7(8).
- Gautam D, Ahmed M, Meena YK, Ul Haq A (2018) Machine learning-based diagnosis of melanoma using macro images. International journal for numerical methods in biomedical engineering 34(5): e2953.
- Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, et al. (2019) End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nature medicine 25: 954-961.
- Bailey MH, Tokheim C, Porta Pardo E, Sengupta S, Bertrand D, et al. (2018) Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 173: 371-385.
- Kandoth C, Mc Lellan MD, Vandin F, Ye K, Niu B, et al. (2013) Mutational landscape and significance across 12 major cancer types. Nature 502: 333-339.
- Leiserson MD, Blokh D, Sharan R, Raphael BJ (2013) Simultaneous identification of multiple driver pathways in cancer. PLoS computational biology 9(5): e1003054.
- Pleasance ED, Cheetham RK, Stephens PJ, Mc Bride DJ, Humphray SJ, et al. (2010) A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191-196.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, et al. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A cancer journal for clinicians 68(6): 394-424.
- Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, et al. (2018) Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer 103: 356-387.
- Thun MJ, DeLancey JO, Center MM, Jemal A, Ward EM (2010) The global burden of cancer: priorities for prevention. Carcinogenesis 31(1): 100-110.
- (2005) World Health Organization. Preventing Chronic Diseases: A Vital Investment, Geneva, Switzerland.
- Rosenthal ET (2011) ASCO, UICC, and ACS Announce Response to Global Cancer Crisis, Urge World Leaders to Act this September. Oncology Times 33: 40-41.
- Hartman AL, Hechtelt Jonker A, Parisi MA, Julkowska D, Lockhart N, et al. (2020) Ethical, legal, and social issues (ELSI) in rare diseases: A landscape analysis from funders. European journal of human genetics: EJHG 28(2): 174-181.
- Mc Ewen JE, Boyer JT, Sun KY, Rothenberg KH, Lockhart NC, et al. (2014) The Ethical, Legal, and Social Implications Program of the National Human Genome Research Institute: Reflections on an ongoing experiment. Annual review of genomics and human genetics 15: 481-505.

 Review Article
Review Article