Abstract
Nucleic acid (DNA) extraction using a Formalin-Fixed Paraffin-Embedded (FFPE) block is a frequent practice in pathological diagnosis and medical research. The extracted nucleic acid, which is the targeted end-product for the further molecular genetic analysis, depends on the ratio of tumor cells to normal cells. Theoretically, the same tissue sample would give an identical genetic detection result. However, the variable percentage of tumor cells and normal cells influences this theoretical result, therefore potentially influencing the final interpretation of this genetic testing result. To investigate the relationship between DNA isolation and biomarker detection, we studied biomarker detection using DNA isolated from FFPE samples and from secondary DNA isolation of leftover tissues that are usually available after the first round of lysis. For this study, primary FFPE samples of malignant pleural mesothelioma (MPM) were used. Genetic p16 (CDKN2A) copy number variation (CNV) analysis was carried out using DNA isolation from the first isolation and from the leftover tissue. We used droplet digital polymerase chain reaction (ddPCR), a precise method to detect CNV. The number of copies of reference genes (E2) were compared with those of p16(CDKN2A), a well-known deleted gene in mesothelioma. Our results indicate that DNA isolated from leftover tissue contains a higher ratio of normal cells compared to tumour cells. This DNA isolation factor can influence the interpretation of genetic alternation detection results.
Keywords: DNA Isolation; ddPCR; p16(CDKN2A); Molecular Biomarker
Introduction
Genetic testing is a type of medical test that identifies changes in chromosomes, genes, or proteins. The results of a genetic test can confirm or rule out a suspected genetic condition or help determine an individual’s chance of developing or passing on a genetic disorder. Genetic testing allows identification of genetic alterations that could also contribute to targeted therapy, such as testing for EGFR and ALK1 in lung cancer [1,2]. ADRI is a research institute whose primary focus is on asbestos related disease especially mesothelioma. Therefore, establishing sound research laboratory techniques including accurate genetic testing and interpretation remains one of our core practices. Conventionally, molecular biomarker detection is carried out by quantitative polymerase chain reaction qPCR, a molecular biotechnology based on the polymerase chain reaction (PCR), that monitors the amplification of a targeted genetic molecule. For this study we have chosen droplet digital PCR (ddPCR). To the best of our knowledge and review of current literature, there is no one single biomarker that can identify mesothelioma. The current biomarkers utilised to detect MPM from adenocarcinoma in clinical diagnosis include CK8/18, Calretinin, CK 5/6, CD141, HBME-1, WT-1, D2-40, EMA, CEA, TAG72, BG8, CD15, TTF-1, and BerEP4. BAP1 and MTAP are recent biomarkers used clinically to help differentiate between reactive mesothelial hyperplasia and MPM.
In our previous study [3], we tested p16(CDKN2A) deletion and BAP1 genomic expression using Fluorescence InSite Hybridization (FISH) testing and CNV analysis with ddPCR respectively [3]. Results indicated that ddPCR provides an advanced biotechnological technique which is a refinement of conventional PCR methods. The direct measurement of DNA and reference allows calculation of CNV using copy number of a reference gene detected by the digital reader of ddPCR. In this study, we used mesothelioma FFPE samples to study p16(CDKN2A) CNV to test our hypothesis that DNA isolation factors can affect the test results. We isolated DNA from FFPE samples and then did a second round of DNA isolation using the leftover tissue from the first isolation. ddPCR was then performed to study CNV.
Materials and Methods
Clinical Samples
11 FFPE samples were included. All samples were approved by the Human Research Ethics Committee at Concord Repatriation General Hospital, Sydney. Written informed consent from all participants were obtained.
DNA Isolation
All procedures were carried out according to manufacturer’s instructions with in-house modification to meet the ISO15189 standard. For this study, QIAamp DNA Mini Kit was used. After first lysis buffer incubation, lysate was used to continue the DNA isolation process. Leftover tissue was then used for the secondround isolation by adding additional lysis buffer then following the isolation process. Both sets of DNA isolation were then quantified by nanophotometer to measure DNA input for ddPCR analysis.
Assay Design
The study design of CVN of p16(CDKN2A) is based on our previous experience from our published paper [3]. To be able to compare copy number precisely, we designed a probe-based assay. We used the sample primer set for p16 from the previous paper [3]. In the same reaction we also designed a p16(CDKN2A) probe labelled with FAM and a reference gene E2 representing two copies from each cell was also included with HEX label. The detected number of E2 copies implies the number of DNA that was analyzed with the designated probe and primer. The number of E2 copies was compared to the number of p16 copies to give a p16:E2 ratio. A p16:E2 ratio approaching 2 is considered no deletion of p16, whilst a p16: E2 ratio tending to 1 is considered to be a heterozygous deletion of p16. When the ratio tends towards 0, this indicates that there is a homozygous deletion of p16.
ddPCR
DNA from the first and second round isolates of FFPE samples were subjected to ddPCR analysis following manufacturer’s instructions for probe assay analysis. Copy number for p16 and E2 were used for CNV calculation. DNA samples were examined by the ddPCR. Droplet digital PCR technology is a digital PCR method utilising a water-oil emulsion droplet system. Droplets are formed in a water-oil emulsion to form the partitions that separate the template DNA molecules. The droplets serve essentially the same function as individual test tubes or wells in a plate in which the PCR reaction takes place, albeit in a much smaller format. The massive sample partitioning is a key aspect of the ddPCR technique. The droplet digital PCR System partitions nucleic acid samples into thousands of nanoliter-sized droplets, and PCR amplification is carried out within each droplet. This technique has a smaller sample requirement than other commercially available digital PCR systems, reducing cost and preserving precious samples.
Results
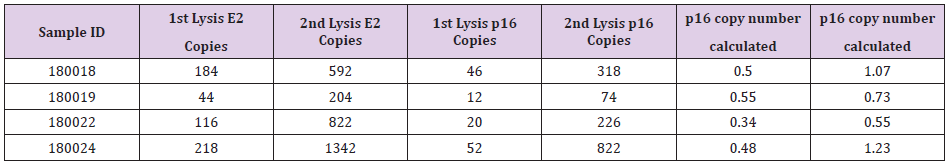
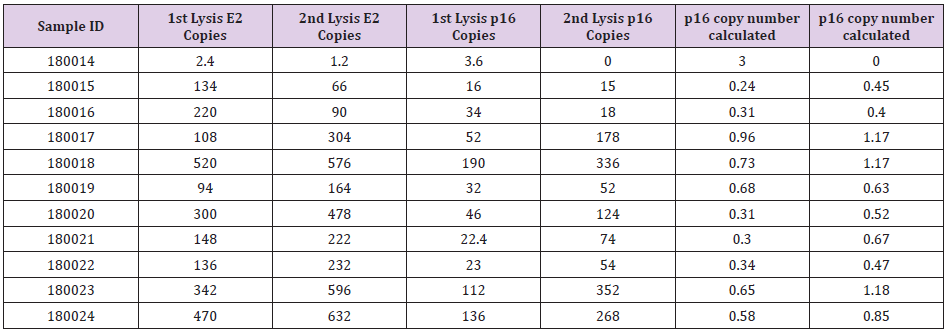
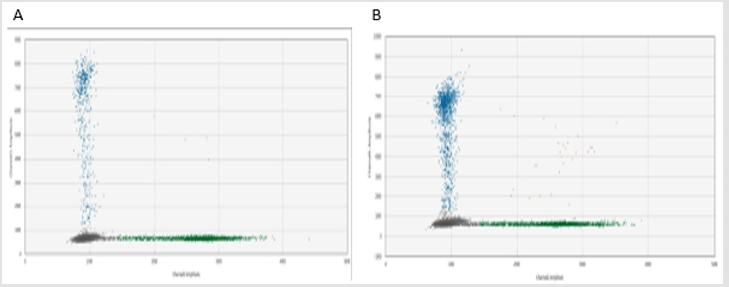
In this study, 11 FFPE samples of MPM patients were included. Qiagen FFPE mini kit was applied to extract DNA from the FFPE scrolls. Two scrolls of 20 μm in thickness were obtained from the FFPE block (numbered as 180014 to 180024 sequentially). DNA concentration was analyzed by Nanophotometer and Nanodrop as reference quality checkpoints. All samples showed good quantity and quality of isolated DNA for ddPCR analysis. All samples were subjected to ddPCR analysis. Raw data was collected and CNV of p16 was calculated as listed in Table 1 (DNA from first isolation) and Table 2 (DNA from second isolation). As indicated in the table, results were obtained for every sample except one (180014). Result for sample 180014 was not comparable. Its DNA content was unusually low which resulted in a large destabilizing influence on the ratio calculation given that the base amount of DNA is effectively absent. This lack of DNA may have been due to the fact that the actual size of primary sample was too small or due to a high fragmentation of DNA in the FFPE block. Figure 1 shows ddPCR results for different populations of E2 (green) and p16 (blue). Copy numbers were then calculated using the CNV calculation formula (copy number detected for p16/copy number of E2) x 2, as every E2 reference refers to two copies in each cell.
The CNV number is then subjected to being called as p16 homozygous or as heterozygous deletion of p16. The same tumour samples from their identical FFPE blocks are expected to give equivalent results. However, we have observed a tendency of higher p16:E2 ratios from the leftover tissue samples compared to the initial samples, which implies that there is a higher number of normal cells in leftover tissue samples. E2 copy was increased in leftover samples compared to the original samples. In all the samples studied in this paper, E2 population was detected as a higher number in all leftover samples (the second DNA isolation) compared to first DNA isolation. The higher E2 copy number in leftover samples influences the final CVN calculation. Samples 180018, 18009, 180022 and 180024 were first carried out by researcher A. E2 copies of these four samples in the leftover tissue showed increase copy number when compared with E2 copies from the first isolations. Researcher B then performed samples 180014 to 180024 (total 11 samples). Except for one sample (180014), all samples also showed elevated E2 copy numbers. Sample 180014 did not show an increased trend potentially due to low DNA quantity from both isolations. We propose that the difference in p16:E2 ratios in first and second DNA isolations may imply that there are more normal cells in the leftover samples resulting in more E2 copies. The results from leftover tissue samples may not represent the actual CNV in clinical samples.
Figure 1: ddPRC analysis, population detected in sample A (first lysis) and B (second lysis) Blue populations detected p16 probe and green populations detected E2 probe.
Discussion and Conclusion
Proteinase is applied to lyse the cells; we presume normal cells have better cellular structure and architecture making them more difficult to break by lysis buffer. Tumour cells lacking in cell-cell adhesion molecules may have low protein content on the cell surface, enhancing the effectiveness of lysis. From this study, we also tried different proteinases and found that proteinase K is better than proteinase as less leftover tissue was retained after lysis step. Our data supports our hypothesis that DNA isolation factors affect CNV results as more E2 copies were detected from the second DNA isolation samples. These results demonstrate the concept that normal lysis of normal cells is more difficult than for the tumour cells, possibly due to the loss of glycoproteins in the tumour MPM cells [4]. We therefore suggest that DNA extracted from leftover tissue samples cannot be used for genetic alternation detection by ddPCR as it may cause the incorrect calculation of CNV of the samples. Our results also demonstrate our hypothesis that given that normal cells are more difficult to lyse, a higher number of normal cells will be retained in the leftover sample, which may induce the problem of false negative results of genetic alternation detection. Since tumour cells have less glycoproteins [4] which are easier to break down during isolation process,we therefore suggest that molecular testing that is focused on tumour cell analysis should only use DNA isolation from first lysis. The data from this study suggest that normal lysis of normal cells is more difficult than for tumour cells and implies the loss of glycoproteins in MPM tumour cells. Further studies will be conducted to investigate the relationship between cell-cell adhesive molecules and the corresponding genes.
Author Contributions
M.L.Y designed and carried out the experiments in this study, draft first version of the manuscript. E.M.R edited and commented on the structure of the manuscript. K.L. is an experienced pathologist that provided professional comments and edited the manuscript, BCM is a cardiothoracic surgeon who provided all necessary biospecimen for this study.
K.H.S carried out experiment of this study. Y.Y.C. conceived the project and drafted the manuscript. All authors approved the submitted version.
Acknowledgment
We would like to acknowledge the support of Prof. Ken Takahashi from the Asbestos Diseases Research Institute for his helpful advice to this work.
References
- Antonicelli A, Cafarotti S, Indini A, Galli A, Russo A, et al. (2013) EGFR-targeted therapy for non-small cell lung cancer: focus on EGFR oncogenic mutation. Int J Med Sci 10(3): 320-330.
- Cheng YY, Rath EM, Linton A, Yuen ML, Takahashi K, et al. (2020) The Current Understanding of Asbestos-Induced Epigenetic Changes Associated with Lung Cancer. Lung Cancer (Auckl) 11: 1-11.
- Sarun KH, Lee K, Williams M, Wright CM, Clarke CJ, et al. (2018) Genomic Deletion of BAP1 and CDKN2A Are Useful Markers for Quality Control of Malignant Pleural Mesothelioma (MPM) Primary Cultures. Int J Mol Sci 19(10).
- Timpe LC, Yen R, Haste NV, Litsakos-Cheung C, Yen TY, et al. (2013) Systemic alteration of cell-surface and secreted glycoprotein expression in malignant breast cancer cell lines. Glycobiology 23(11): 1240-1249.

 Short Communication
Short Communication