Abstract
Background:
Abbreviations: CRC: Colorectal Cancer; HNPCC: Hereditary Non-Polyposis Colorectal Cancer; FAP: Familial Adenomatous Polyposis; MAP: MUTYH-Associated Polyposis; NGS: Next Generation Sequencing
Opinion
The introduction of Next Generation Sequencing (NGS) technology in clinical practice has had a profound impact in the identification of gene variants responsible for heritable diseases. The high throughput capability of NGS allows the simultaneous screening of the whole coding genome (usually defined clinical exome) at affordable costs and with reasonable turnaround times. While the traditional single gene sequencing approach had low detection power, NGS greatly expanded the number of tested genes in many genetic conditions where disease predisposition can be due to causative mutations in many different genetic loci. In Colorectal Cancer (CRC), which account for 10% of all newly diagnosed cancers and is responsible for 9% of all cancer deaths, family history is a major risk factor [1-3]. It is estimated that approximately a quarter of all CRCs is due to familial inheritance with 5-8% of cases attributable to single gene inherited predisposition [4,5]. Traditionally, three major types of inherited predisposition to CRC are recognized being the Lynch syndrome (previously known as Hereditary Non-Polyposis Colorectal Cancer or HNPCC), Familial Adenomatous Polyposis (FAP), and the MUTYH-Associated Polyposis (MAP).
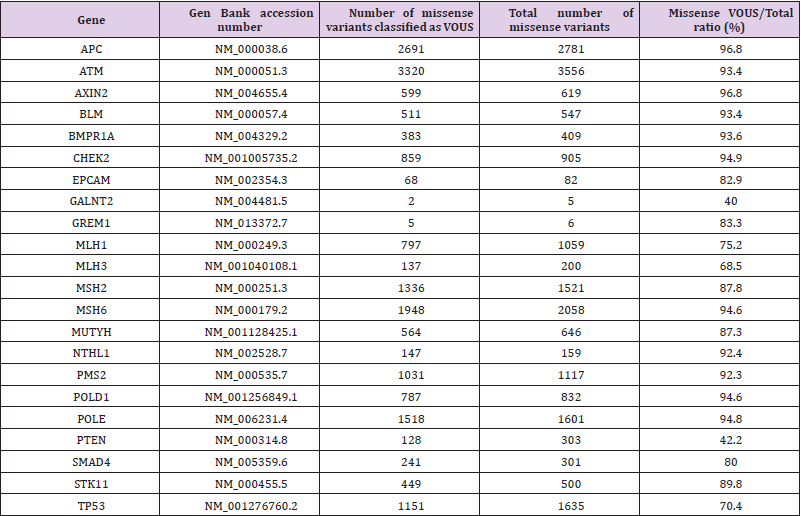
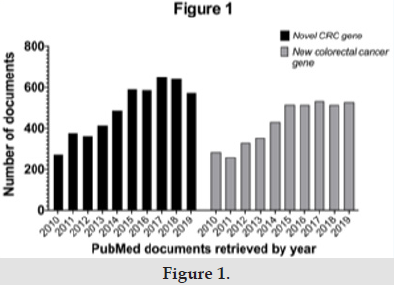
These three CRC-predisposing conditions are mainly caused by pathogenic variants in six primary genes (MLH1, MSH2, MSH6, PMS2, APC, and MUTYH). However, several additional genes are also known to cause hereditary CRCs or conferring an increased risk for colorectal cancer. Thanks to NGS technology, the labor extensive and time-consuming Sanger sequencing approach has been replaced by multigene panels that allow for an inexpensive and comprehensive screening for numerous hereditary CRC genes. Yet, NGS has been less successful in identifying novel CRC-predisposing genes [6,7]. Indeed, a PubMed search (accessed on February 5th 2020) using the terms “new colorectal cancer gene” and “novel colorectal cancer gene”, reveals that documents retrieved were constantly increasing in the five years period 2010-2014, while remained steady in the following 2015-2019 period (Figure 1). The National Cancer Comprehensive Network (NCCN), has recently recommended a list of 22 genes to be included in multigene testing panel for hereditary CRCs [8]. However, for all the 22 genes, the missense variants listed at Varsome (accessed 30th January 2020), and classified as of unknown significance (VOUS), largely outnumber variants classified as pathogenic or likely pathogenic (Table 1).
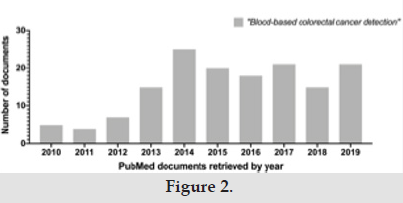
Thus, it is possible that the missing heritability of familial colorectal cancer could be explained by a re-evaluation of the putative pathogenic role of VOUS, at least in some families, rather than attributable to yet unknown predisposing genes. Recent work has demonstrated the utility of machine learning training in decreasing the number of VOUS in different disease-causing genes [9,10]. Similar approaches could be useful also applied to hereditary cancer syndromes. Another unresolved issue in hereditary predisposition to CRCs is the lack of sensitive and reliable bloodbased tests to diagnose cancer in its early preclinical stages that would be pivotal in reducing the cancer burden in patients at high risk. Though, like the search for novel CRC predisposing genes, the number of PubMed documents retrieved using the search term “blood-based colorectal cancer detection” raised constantly in the 2010-2014 period while plateauing in the following five years (2015-2019, Figure 2). Despite the huge emphasis that the concept of “liquid biopsy” has provoked in cancer screening as an attractive tool for early detection and minimally invasive diagnosis of cancer, the sensitivity and the specificity achieved so far prevented these tests to be used in general population screening (reviewed in [11- 15]).
The liquid biopsy approach has been focused on analyzing either Circulating Tumor Cells (CTCs), cell-free circulating tumor DNA (ctDNA) and/or tumor-derived extracellular vesicles (exosomes). Liquid biopsy approaches have been proven extremely useful in monitoring cancer recurrence, progression, efficacy of therapeutic intervention as well as disease profiling, considering the clonal heterogeneity of human cancers [16,17]. Blood-based screening methods were less effective in detection of cancer in early preclinical stages. However, a recent study demonstrated that using a machine learning approach it is possible to achieve high sensitivity and specificity applying whole-genome sequencing of cell-free DNA on a cohort of predominantly early stage CRCs [18]. In this light, tumors exhibiting Microsatellite Instability (MSI), may represent a promising target for future applications. In fact, several recent works have determined that the MSI phenotype may be detected directly from cell-free DNA [19-23]. In conclusion, recent advances should help in a more effective monitoring of patients with high CRC risk and the long-term surveillance of their health status.
Declaration of Conflict Interest
The author declares no potential conflict of interest with respect to the research, authorship, and/or publications of this article
Funding
This work was supported by a grant from Fondazione Puglia entitled “Ricerca di nuovi geni di predisposizione e di markers predittivi di neoplasia nelle sindromi di predisposizione ereditaria al cancro del colon retto”.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6): 394-424.
- Markowitz SD, Bertagnolli MM (2009) Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med 361(25): 2449-2460.
- Frank C, Fallah M, Sundquist J, Hemminki A, Hemminki K (2015) Population Landscape of Familial Cancer. Sci Rep 5: 12891.
- Lu C, Xie M, Wendl MC (2015) Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun 6: 10086.
- Huang KL, Mashl RJ, Wu Y (2018) Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 173(2): 355-370.
- Campbell BB, Light N, Fabrizio D (2017) Comprehensive Analysis of Hypermutation in Human Cancer. Cell 171(5): 1042-1056
- Grolleman JE, de Voer RM, Elsayed FA (2019) Mutational Signature Analysis Reveals NTHL1 Deficiency to Cause a Multi-tumor Phenotype. Cancer Cell 35(2): 256-266.
- (2018) National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for genetic/familial high-risk assessment: colorectal V.1.
- Accetturo M, D Uggento AM, Portincasa P, Stella A (2019) Improvement of MEFV gene variants classification to aid treatment decision making in familial Mediterranean fever Rheumatology (Oxford)
- Accetturo M, Bartolomeo N, Stella A (2020) In-silico Analysis of NF1 Missense Variants in ClinVar: Translating Variant Predictions into Variant Interpretation and Classification. Int J Mol Sci 21(3): E721.
- Finotti A, Allegretti M, Gasparello J (2018) Liquid biopsy and PCR-free ultrasensitive detection systems in oncology (Review). Int J Oncol 53(4): 1395-1434.
- Marrugo Ramirez J, Mir M, Samitier J (2018) Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int J Mol Sci 19(10): 2877
- Massihnia D, Pizzutilo EG, Amatu A (2019) Liquid biopsy for rectal cancer: A systematic review. Cancer Treat Rev 79: 101893.
- Mathai RA, Vidya RVS, Reddy BS (2019) Potential Utility of Liquid Biopsy as a Diagnostic and Prognostic Tool for the Assessment of Solid Tumors: Implications in the Precision Oncology. J Clin Med 8(3): 373.
- Zhang W, Xia W, Lv Z, Ni C, Xin Y (2017) Liquid Biopsy for Cancer: Circulating Tumor Cells, Circulating Free DNA or Exosomes? Cell. Physiol. Biochem 41(2): 755-768.
- Schröck A, Leisse A, de Vos L, Gevensleben H, Dröge F, et al. (2017) Free-circulating methylated DNA in blood for diagnosis, staging, prognosis, and monitoring of head and neck squamous cell carcinoma patients: An observational prospective cohort study. Clin Chem 63(7): 1288-1296.
- Quandt D, Dieter Zucht H, Amann A, Wulf Goldenberg A, Borrebaeck C, et al. (2018) Implementing liquid biopsies into clinical decision making for cancer immunotherapy. Oncotarget 8(29): 48507-48520.
- Wan N, Weinberg D, Liu TY (2019) Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer 19(1): 832.
- Sun J, Fei F, Zhang M (2019) The role of mSEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer 19(1): 450.
- (2019) SILAC-Based Quantification of TGFBR2-Regulated Protein Expression in Extracellular Vesicles of Microsatellite Unstable Colorectal Cancers. Int J Mol Sci 20(17): 4162.
- Georgiadis A, Durham JN, Keefer LA (2019) Noninvasive Detection of Microsatellite Instability and High Tumor Mutation Burden in Cancer Patients Treated with PD-1 Blockade. Clin Cancer Res 25: 7024-7034.
- Willis J, Lefterova MI, Artyomenko A (2019) Validation of Microsatellite Instability Detection Using a Comprehensive Plasma-Based Genotyping Panel. Clin Cancer Res 25(23): 7035-7045.
- Mayrhofer M, De Laere B, Whitington T (2018) Cell-free DNA profiling of metastatic prostate cancer reveals microsatellite instability, structural rearrangements and clonal hematopoiesis. Genome Med 10(1): 85.

 Opinion
Opinion