Abstract
The gut microbiota is a diverse system with important roles in host functions and well-being, and its dysbiosis is linked to metabolic disorders like obesity and diabetes. Stress can affect microbiota population and its interaction with the host. Humanized animal models created by transplanting human microbiota into germ-free (GF) animals have been used to study human-derived microbiota population dynamics, and the relationship between microbiota dysbiosis and altered host functions. A limited number of GF variants are available commercially, and humanized models generated through mono-association are donor-specific that may not represent the entire spectrum of human microbiota. To address these issues, we developed a new Depersonalized Humanized Rat (DHR) Model that harbors a homogeneous profile of human microbiota derived from multiple donors. Initially, pseudo germ-free (PGF) Sprague Dawley rats were produced, using an antibiotic cocktail. These PGF rats were subsequently transplanted with the fecal materials from multiple human donors. Elimination of donor-specific characteristics was achieved by coprophagic transfer of the transplanted microbiota between humanized rats. Metagenomic analysis confirmed the emergence of a relatively homogenous profile of human microbiota after 5 weeks, which was stable up to 15 weeks. The analysis of HPA hormones, proinflammatory cytokines and other markers did not reveal any significant changes in the humanized rats. Thus, procedures based on this approach may be used to generate a donor-independent, humanized model from different native or modified rat strains, which in turn could greatly facilitate future studies of human microbiota and its effects on host functions.
Keywords: Antibiotic Cocktail; Coprophagic; Depersonalization; DHR Model; Fecal Transplant; Gut Microbiota; Humanization; 16s rRNA Hypervariable Regions; Pseudo- Germ Free; Sequencing Analysis
Introduction
The human microbiota is a complex and diverse system with great interpersonal variability that is affected by genetics, lifestyle, diet, health, and geography [1]. It has many functions such as maintaining and preventing injury to intestinal epithelial cells through interaction with toll-like receptors [2] and regulating fat storage and metabolism to provide energy for colonic epithelial cells [3].The importance of gut microbiota in metabolic disorders like obesity and diabetes has also been demonstrated [4]. However studying the gut microbiota in human subjects is technically challenging, because there is no reliable, non-invasive method for collecting region-specific samples of the gut microbiota. Humanized models generated by transplanting human microbiota to germfree (GF) animals are useful for studying the changes in microbial population structure and functions following stress exposure, as well as the relationship between microbiota dysbiosis and changes in host functions [5,6], especially when the analysis requires tissue samples from the hosts [7,8]. GF models from commercial venders, however, are limited and GF rats are not commercially available in the United States. Establishing and maintaining a GF colony in-house is costly, due to the requirement of specialized facilities. Additionally, humanized models, generated by mono-association with a single human donor, compound the variability that already exists due to inter-individual differences in gut microbiota, resulting in inconsistencies in the response of microbiota to experimental manipulation [9]. One way to overcome this challenge is to generate multiple humanized lines and use them as biological replicates; however, this greatly increases the cost and the number of animals needed to account properly for such variability.
The goal of this study was thus to develop a reliable and reproducible method for generating pseudo germ-free (PGF) rats that are suitable for the production of humanized rats harboring a stable, homogenous, gut microbiota derived from multiple human donors. Others have described the use of antibiotics to deplete the native gut microbiota of rodents [9-13].The type of antibiotics used and method of delivery can affect the health of the animals [14] and the diversity of the remaining gut microbiota that can survive the treatment [11]. Although the microbiota that colonize the gut of GF animals after the transplant of human fecal microbiota showed similarity to the donor material, previous reports showed that the microbial population re-established in antibiotic-treated animals after fecal transplant has greater similarity with the native microbiota [12,13] highlighting the need to improve the procedures for generating PGF and humanized models. We report here the development of a new antibiotic cocktail, delivered in the drinking water. This cocktail causes minimal systemic exposure, thereby avoiding potential adverse effects on host functions. It was optimized with the lowest concentration and type of antibiotics while capable of significantly decimating the rat native gut microbiota, thus generating PGF rats.
Humanization was accomplished using repeated inoculations of fecal microbiota from human donors, which has been successfully employed by others [9]; however, this study took advantage of coprophagia, the natural rodent behavior of consuming fecal materials, for the horizontal transfer and exchange of the transplanted human-derived microbiota between animals that do not share the same housing cage. This approach effectively increased the number of inoculations and further blended the microbiota across recipients, establishing a homogenous human microbiota population within the rat community. The homogeneity and stability of the transplanted microbiota was confirmed by metagenomic analysis targeting the hypervariable region of the 16S rRNA locus in the microbiota genomic DNA.
This procedure is capable of stably establishing human microbiota in antibiotic-generated PGF rats and eliminated donor-specific characteristics in the humanized rats. The resultant model is thus termed Depersonalized Humanized Rat (DHR) Model. This model should be a valuable tool for studying the effects of dysbiosis of human gut microbiota and the interaction between the human microbiota and the host functions under dietary imbalance, disease states or other physiological stressors such as sleep deprivation and circadian dyssynchrony.
Animals, Materials and Methods
Animals and Husbandry
The study protocol was approved by the Wright-Patterson Air Force Base Institute of Research, Institutional Animal Care and Use Committee (IACUC), and the U.S. Air Force Surgeon General’s Office of Research Oversight and Compliance. The experiments reported herein were conducted in compliance with the Animal Welfare Act and in accordance with the principles set forth in the “Guide for the Care and Use of Laboratory Animals,” Institute of Laboratory Animal Research, National Research Council, National Academies Press, 2011, and in a facility accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). The animals were Specific Pathogen Free, Male, Sprague Dawley (SD) Rats (Rattus Norvegicus) (Charles River Laboratories, Wilmington, MA), CRL:CD(SD), which have been successfully employed by others in humanization models. Animals were 6 weeks old at study initiation. Only males were used to avoid sexspecific interactions that might compound results during model development. The rats were socially housed (2/cage) in clear plastic cages and provided tunnels, nesting material, and nylon bones for enrichment. naïve and normal cage control animals had conventional bedding (Cell Zorb, Cincinnati Lab Supply, Cincinnati, OH) and food (Lab Diet Formulab Diet 5008, Cincinnati Lab Supply), and water were freely available. Animals in the sterile environment were similarly housed but the cages and enrichment objects were sterilized, and sterile food (Picolab Rodent Irradiated Chow, Charles River), bedding (Alpha Dri Irradiated Bedding, Charles River), and autoclaved water were used. The animal rooms were climate controlled (20 – 26 °C, 30 – 70% humidity) with 12-hour light/dark cycle (on at 0600).
Animals were examined and weighed upon arrival and placed under quarantine pending the results of representative blood tests for 10 specific rodent pathogens. Assignment of animals to experimental groups was achieved using a random number generator. All animals were examined visually at least twice daily throughout the study for signs of distress.
Microbial Titer Determination
Sterile 1x phosphate buffered saline, pH 7.3 (PBS) (Thermo Fisher Scientific (Fisher), Carlsbad, CA) was used for dilution. Fecal samples were homogenized using a sterile, mini pestle and dissolved in 0.75 mL of PBS. Tissue samples (including, small intestine, cecum, large intestine, mesentery, lymph nodes, spleen, and liver) were collected post-mortem, flash frozen in liquid nitrogen and stored at -80 °C until use. The frozen tissues were later homogenized in 0.5 ml of PBS using the Bullet Blender Gold (2 minutes on speed 6), RhinoVials, and 2.0 mm Zirconium Oxide Beads (3/ tube) (all from Next Advance, Troy, NY). A 0.25-mL portion of the homogenate (with appropriate dilutions when necessary) was spread evenly onto blood-agar plates (tryptone soya agar containing 5% sheep blood, Fisher) in duplicate. Plates were incubated at 37 °C overnight in a standard incubator or using the BD GasPak EZ Gas Generating System (Fisher) for anaerobic growth. The colonies were enumerated by the Scan 300 Colony Counter (Interscience Laboratories, Woburn, MA) and expressed as total (aerobic + anaerobic) colony forming unit (CFU) per gram of material.
Development of Antibiotic Cocktail
Initially, Enrofloxacin (500 mg/L), Neomycin (150 mg/L), Vancomycin (350 mg/L), Amphotericin B (1000 mg/L), and Ampicillin (150 mg/L) (all from Patterson Vet, Inc.) were chosen as candidates because of their low gastrointestinal absorption or rapid elimination thereby minimizing systemic exposure and residual effects. The individual and combined effectiveness of these agents were tested using plate cultures to determine their effects on microbial titer of the cecal contents from naïve rats as described above but while adding the antibiotic(s) to the blood-agar plates. Amphotericin B and Ampicillin, which did not show significant antimicrobial activity when used alone, nor significantly improved the cocktail’s effectiveness, were eliminated after the first round of testing. The concentrations of remaining antibiotics were adjusted empirically until a combination that effectively resulted in no detectable colony on agar plates was discovered.
Generating Pseudo Germ-Free (PGF) Rats
The animals were housed in Semi-Rigid GF Isolators (Charles River Laboratories) that were maintained in the PGF animal production room following the manufacturer’s recommendations. The GF status of the isolators was monitored using culture swabs and Pocket Swab Plus (Charm Sciences, Lawrence, MA). After two days of acclimation to the isolators, the rat native microbiota was depleted using the antibiotic cocktail in the drinking water, which was replaced daily for 14 days. Untreated rats, housed in a separate isolator and receiving regular food and drinking water, were used as control. Tests indicated that cecal content was a more reliable indicator than feces for the evaluation of the depletion of microbiota in the gut. Thus, the bacterial titer of the cecal contents was determined in two animals at each of Days 3, 6, 9 and 12 (Day 1 = the first day of antibiotics treatment), in order to monitor the depletion of the native rat microbiota. Initially, the antibiotic cocktail optimized using the plate culture method (65 mg/L Enrofloxacin, 1000 mg/L Neomycin, 500 mg/L Vancomycin) was used. The concentration of antibiotics were further optimized empirically until no detectable microbial colony (using the plate culture method) in the cecal content after three days of antibiotic treatment.
Human Fecal Microbiota
Fecal samples from normal, healthy, human donors were supplied by OpenBiome (Somerville, MA) through a contractual agreement. The subjects were Caucasian males aged 20-45, a body mass index of 19-25, no drug abuse, no medications including antibiotics for at least 12 months, no infections, and no gastrointestinal, immunosuppressive, autoimmune, psychological, neurological, or sleep disorders. Fecal samples from six human donors were selected and used in his study.
Depersonalized Humanized Rat (DHR) Model Development
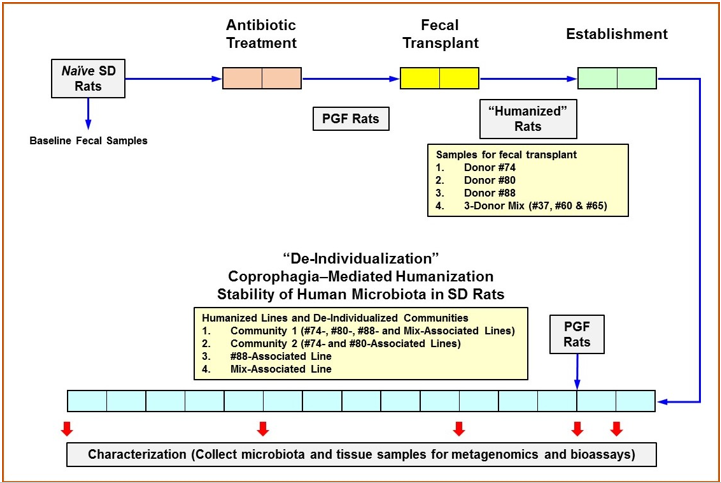
The experimental design of DHR Model development is illustrated in Figure 1. This 4-step procedure lasted a total of 21 weeks: 2 weeks each for Antibiotic Treatment, Fecal Transplant, and Establishment; and 15 weeks for the evaluation of the depersonalization process. During antibiotic treatment of rats, fecal samples were collected daily, and 2 rats were euthanized at Days 5 and 10 for the measurement of the microbial titer of the fecal samples and cecal contents, respectively. Fecal transplant was accomplished by intra-gastric gavage with 8.5 x 108 CFU of human fecal microbiota suspended in 0.5 mL of 1x lysogeny broth (LB) (Fisher). The inoculum was derived either from a single donor (donors #74, #80 or #88) or from a mix of equal contributions from donors #37, #60 and #65 (3-donor mix for multi-association). Each PGF rat received five inoculations, one each at Days 15, 17, 19, 22, and 25. During the two weeks of establishment, the microbial titer was determined from daily fecal samples and cecal samples collected at Days 26 and 40 (2 rats for each timepoint) to confirm the colonization of the transplanted human microbiota in the rat hosts.
Depersonalization of humanized rats began by forming two communities. Community 1 was comprised of half the animals from each of the following humanized lines: #74-associated, #80-associated, #88-associated, and 3-donor-mix-associated. Community 2 was comprised of the remaining half of the #74- and #80-associated lines. The remaining half of the #88- and 3-donormix- associated animals were maintained as independent lines. Each community was depersonalized by coprophagia-mediated horizontal transfer of the gut microbiota within each community. On every second day for 15 weeks, the bedding including all fecal pellets from all members of the respective community was pooled, mixed with fresh bedding, and redistributed back to the members’ cages. This is to allow all members of the community had the opportunity to consume each other’s feces, even though they did not share the same cage. Each community was housed in a separate isolator to prevent cross-contamination or the introduction of external microbes. Normal rats (with no antibiotic treatment or fecal transplant), housed in a separate isolator and receiving regular food and drinking water, were used as control. The acquisition of human microbiota through coprophagia alone was also tested by introducing newly generated PGF rats to the communities and providing them with a portion of the mixed bedding that contains fecal pellets from humanized rats. Up to four animals per community were euthanized at Weeks 1, 2, 5, 10 and 15 of the Depersonalization period to determine the titer and population structure of the gut microbiota. Markers of selected host functions were also monitored. Lipopolysaccharide binding protein (LBP) and myeloperoxidase (MPO) activity were determined using serum and intestinal tissues, respectively, to detect changes in the intestinal barrier function. Peripheral corticotrophin releasing hormone (pCRF), adrenocorticotropic hormone (ACTH), and corticosterone were measured in serum to detect changes associated with the hypothalamus-pituitary-adrenal (HPA) axis. Serum levels of a panel of proinflammatory cytokines were quantified to assess inflammatory response in the humanized animals. The bioassay methods and results are described in the supplemental data.
Figure 1: Experimental Design.
Note: Each box represents 1 week. The native gut microbiota of Sprague Dawley (SD) rats was depleted by antibiotics treatment in germ-free isolators to produce pseudo germ-free (PGF) rats. Humanization was accomplished with five (5) intragastric inoculations of human fecal material followed by 2 weeks of establishment for the colonization of the transplanted human microbiota. Some rats receiving fecal transplant were maintained as individual lines while others were assigned to specific communities for coprophagia-mediated depersonalization. Community bedding containing fecal pellets was mixed and redistributed to promote the horizontal transfer of microbiota. Fecal and cecal microbiota, as well as tissues samples were collected at indicated timepoints for 16s rRNA hypervariable regions sequencing analysis and the analysis of various markers for host functions.
16s rRNA Hypervariable Regions Sequencing Analysis of Gut Microbiota
The population structure of the gut microbiota was analyzed using 16s rRNA hypervariable regions sequencing. Fecal and cecal samples were collected, flash frozen in liquid nitrogen and stored at -80 °C until use. Total DNA was isolated from 250 mg of each sample using the QIAamp DNA Stool Mini Kit (Qiagen, Carlsbad, CA). DNA was eluted in 0.15 mL of AE elution buffer (Qiagen), quantified using the ND-1000 NanoDrop Spectrophotomer (Fisher), and stored at -20 °C until amplified. Hypervariable regions of 16S rDNA (including negative and positive controls) were amplified using the Ion 16S Metagenomics Kit (Fisher). This kit contains two wide-ranging primer sets (V2-4-8 and V3- 6, 7-9) for multiple hypervariable regions, allowing detection of a broad range of microbial species. DNA enrichment after PCR was confirmed by electrophoresis in a 2% agarose gel. PCR products was purified using Agencourt AMPure XP magnetic beads (Fisher) and quantified on the 4200 Tape Station using D1000 screen tape and reagents (Agilent Technologies, Santa Clara, CA). The DNA was sheared enzymatically with the Ion Plus Fragment Library Kit (Life Technologies, Waltham, MA). Samples were labeled individually to allow for multiplexing on the Ion Xpress Barcode Adapters 1-96 Kit (Fisher). For metagenomic sequencing and taxonomic analysis, each DNA library was diluted to 60 pM and pooled. Amplification by emulsion PCR, template preparation, and chip loading were automated using the Ion Chef System (Life Technologies) utilizing Ion 530 chips/kits (Fisher). DNA was sequenced using the Ion Torrent S5 Semiconductor Sequencer v5.2 and Ion S5 Sequencing Kit (Fisher). The metagenomic library was constructed with 400- bp inserts for each sample. Quantitative Insights into Microbial Ecology (QIIME) software (v1.9.1) [14,15] was used for taxonomic analysis of the raw sequences. Operational taxonomic units (OTUs) were assigned based on 97% sequence identity to the Green Genes reference (v13.5) [16]. Beta-diversity was calculated using UniFrac [17] methodology, and three-dimensional principal coordinate analysis (PCoA) plots visualized with Emperor [18].
Result
Development of Pseudo Germ-Free (PGF) Rats
The antibiotic cocktail developed using the plate culture method failed to eliminate all culturable microbial cells in the cecal content from the treated rats. After several rounds of optimization of the antibiotic concentrations, the finalized cocktail (200 mg/L Enrofloxacin, 800 mg/L Neomycin, and 850 mg/L Vancomycin) was able to eliminate all culturable cells in the cecal content after 3 days. Comparing to two to three thousands of colonies normally obtained from the cecal content of control animals (diluted 106-fold), this antibiotic cocktail is able to reduce the microbial load in the cecum (and perhaps in the entire intestinal tract) by at least 3-order of magnitude in 3 days. Based on this result, it was concluded that the treated animals likely reached a PGF status after the course of 14-day antibiotics treatment. However, since a significant portion of the gut microbiota is not culturable, this result may not provide a complete picture concerning the overall effectiveness of this antibiotic cocktail. We found that PCR products of the bacterial 16s rRNA gene hypervariable regions remained detectable in the fecal content from the animals that have received 12-day antibiotics treatment, provided the entire PCR product was used in gel electrophoresis. This result was not completely unexpected, since PCR is highly sensitive that it can detect a few molecules under optimal conditions. For instance, genomic DNA from non-culturable (or even non-proliferative) microbes would give positive results in PCR assays. Consistent with this, the result of metagenomic analysis showed that this antibiotic cocktail resulted in a 52.3% reduction in the number of OTUs detectable in the fecal content.
Microbiota-Humanization of PGF Rats
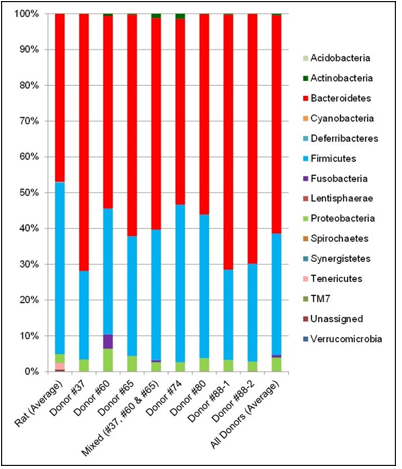
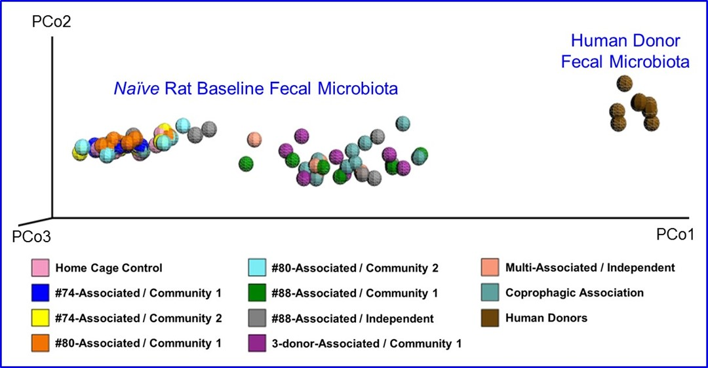
The fecal microbiota from six human donors and naïve rats (prior to antibiotic treatment) were analyzed using 16s rRNA hypervariable regions sequencing to determine their overall diversity and the relative abundancies of various bacterial taxa. The relative abundancies of Bacteroidetes and Firmicutes, the most abundant phyla in Western human populations [19], are quite diverse among the donors recruited for this study. As shown in Table 1, Bacteroidetes ranged from 51.9–71.8% and Firmicutes from 24.7–44.0%, which is consistent with other reports in the literature. [17,20]. The human microbiota is notably different from that from naïve rats (Figure 2). In addition, other phyla including Actinobacteria, Fusobacteria, Proteobacteria, and several minor phyla such as Cyanobacteria, Deferribacteres, Synergistetes, TM7, Tenericutes and Verrucomicrobia were also detected. Donor #60 had a higher level of Fusobacteria than other donors.Some species of Fusobacteria are associated with human diseases [21], but the rigorous donor screening did not reveal signs of infection or other health issues in this donor. Thus, Donor #60 was retained in this study. PCoA plots of the UniFrac distance metric were composed using Emperor, in order to investigate the population structure of the human fecal microbiota to that of the naïve rats. The result of this analysis showed that the human samples formed one cluster well separated from the naïve rat samples (Figure 3).
Table 1: Relative Abundance of Bacteroidetes and Firmicutes in the Fecal Microbiota from Human Donors and Naïve Sprague Dawley Rats.
Note: The fecal microbiota from six human donors, a mixed sample from 3 donors, and naïve rats were analyzed by 16s rRNA hypervariable regions sequencing, and the relative abundance of the phyla Bacteroidetes and Firmicutes are shown. Two independent results (from 2 different fecal donations) are presented for Donor #88.
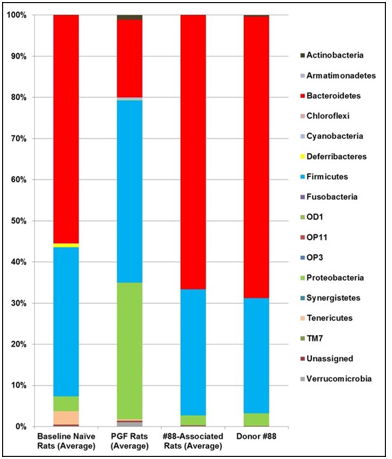
The optimal titer and procedure for human fecal transplant were determined using several pilot experiments. The successful transfer of human microbiota required at least three inoculations beginning immediately after antibiotic treatment. The finalized procedure included one inoculation on each of five days (Days 15, 17, 19, 22, and 25) after the 14-day antibiotics treatment. The colonization of the human-derived microbiota in the rat hosts was monitored after 2 weeks of establishment following the final inoculation. Consistent with previous result, the fecal microbiota from naïve rats was notably different from that of the human donors (see Figure 4, with Donor #88 as a representative example). Interestingly, antibiotics treatment of rats altered their microbial population, which is significantly different from that of the naïve rats or the human donor. However, the transplantation of human fecal material into the PGF rats resulted in a microbiota profile that is virtually indistinguishable from the that of the human donor (Figure 4, #88-Associated Rats compared to Donor #88), thus confirming that this humanization procedure was highly effective in establishing the human microbiota in the PGF rats.
Figure 2: Taxa Bar Plot of Fecal Microbiota from naïve Rats and Human Donors.
Note: The relative abundancies of detectable bacterial phyla in fecal samples were determined by 16s rRNA hypervariable regions sequencing analysis and are displayed here as the average value across all naïve rats (i.e. prior to antibiotics treatment), average value across all human donors, individual donors, and the 3-donor blend used for the mix-associated line. Two independent test results (from two different fecal donations) are shown for Donor #88.
Figure 3: Principal Coordinate Analysis of Microbial Diversity in Fecal Samples from naïve Rats and Humans Donors at Baseline.
Note: Fecal samples were analyzed by 16s rRNA hypervariable regions sequencing to generate a principal coordinate analysis (PCoA) plot comparing the gut microbiota from naïve rats and human donors. Samples from naïve rats are labeled with the human donor and depersonalization community or independent line to which they were assigned. Coprophogic Association are samples from rats assigned to human microbiota exposure only through coprophagia-mediated horizontal transfer in their respective community.
Figure 4: Taxa Bar Plot of the Fecal Microbiota from naïve Rats, Pseudo Germ-Free Rats, Humanized Rats and Human Donor #88.
Note: The relative abundancies of detectable bacterial phyla in fecal samples were determined by 16s rRNA hypervariable regions sequencing analysis. Compared here are the average relative abundancies in the native rat microbiota collected at baseline, samples from PGF rats collected after 12-day antibiotic treatment, samples from #88-associated humanized line collected prior to depersonalization, and that from Donor #88.
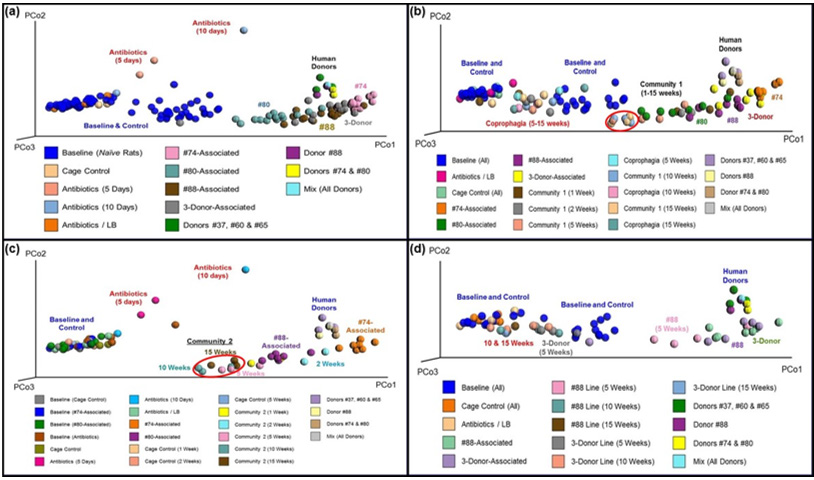
Depersonalization of Humanized Rats
Elimination of the donor-specific characteristics of the gut microbiota in the humanized rats was accomplished through coprophagia, and its progress was monitored using 16s rRNA hypervariable regions sequencing analysis of the fecal samples collected at specific timepoints during the Depersonalization period. Figure 5 displays four PCoA plots of the UniFrac distance metric between fecal samples collected from different experimental groups and human donors. Prior to depersonalization (Figure 5a), all four humanized lines (#74-, #80-, #88-, and 3-donor-mix-associated) formed somewhat overlapping clusters that were distinct from the samples of naïve and control rats. Of the four humanized lines, #74-associated rats formed a tight cluster while those of the other three humanized lines were less compact. Most of the humanized rats (except a couple from the #80-humanized line and one from the mix-humanized line) are close to the human donors and far away from the naïve rat samples. This result shows that the gut microbiota from humanized rats has greater similarity to the human microbiota than to the native rat gut microbiota, thus confirming the successful colonization of the rat intestinal tract by the human microbiota.
The rats in Community 1 appear to have maintained human microbiota throughout the entire 15-week Depersonalization period (Figure 5b). The samples collected at 10 and 15 weeks form a tight cluster (red oval in Figure 5b), indicating that a relatively homogenous profile of microbiota emerged in Community 1 by week 10. The gut microbiota of this community, however, appeared to gradually shift toward the naïve rat cluster, suggesting there may be some degree of instability in the transplanted microbiota over time. Figure 5b also shows that humanization of PGF rats through coprophagia alone (i.e., without direct inoculation of human fecal material) was not successful, since all of the samples from that group were closed to the naïve rat clusters. The rats in Community 2 showed a similar pattern (Figure 5c), with the homogenous profile of the gut microbiota emerged early at the 5-week timepoint and maintained until the end of the experiment (red oval in Figure 5c). The faster depersonalization in Community 2 likely because it had to blend fewer individualized microbial populations. While Community 1 utilized all four humanized lines developed from a total of six human donors, Community 2 was composed of only two humanized lines developed from 2 individual human donors. Similar to Community 1, this community also drifted toward the clusters of rat baseline and cage control samples, suggesting a gradual decrease in the similarity to the human microbiota. Nevertheless, Community 2 remained well separated from the baseline and control rat clusters, even after 15 weeks. These results further confirmed that human microbiota can be successfully established in the PGF rats that was generated using our newly developed antibiotic cocktail and maintained for at least 15 weeks. Moreover, the donor-specific microbiota profile can be eliminated through the sharing of bedding material and fecal pellets.
Figure 5: Principal Coordinate Analysis of Fecal Microbiota from Humanized Rats.
Note: Principal coordinate analysis (PCoA) plots were generated from the UniFrac distance metric using fecal samples from humanized rats and human donors at specific timepoints. Samples collected from human-associated rats at Day 12 of the Establishment period (panel a). Samples collected from Community 1 (panel b) or Community 2 (panel c) at Weeks 1, 2, 5, 10 and 15 of the Depersonalization periods; red ovals depict a relative homogeneous community microbiota. Samples collected from #88-associated and 3-Donor-Mix-associated lines at Weeks 5, 10 and 15 of the Depersonalization periods (panel d). In each panel, cage control represents normal rats with no antibiotic’s treatment or fecal transplant. Antibiotic/LB rats were treated with antibiotics but not human fecal material. The “3-Donor-associated” and “3-Donor-Line” are rats that were transplanted with the mix of materials from Donors #37, #60 and #65. “Donors” indicates fecal samples from individual donor or a mixture of the indicated human donors.
It was quite unexpected that the human microbiota was less stable in rat hosts when maintained as individual lines when compared to the two depersonalized communities. As shown in Figure 5d, only the 5-week samples from the #88-associated line remained separated from the naïve rat cluster. The samples collected at 10 and 15 weeks were closed to the naïve and control rat samples, indicating re-emerging of the native rat microbiota in these humanized animals. Similarly, the 3-donor mix-associated line also appeared to have reverted to the native rat microbiota by week 5. Analysis of markers related to HPA axis, immune, and intestine mucosal functions in the hosts showed that there were no significant changes in these parameters (see Supplemental Data). These results thus suggest that the human microbiota likely interacts with the rat intestinal tract in a similar way as the rat native microbiota, and that the procedure presented here for developing the DHR Model is unlikely to cause any significant change in the host functions.
Discussion
In this study, naïve SD rats were first converted to PGF rats using a highly effective antibiotic cocktail that can deplete all culturable microbial cells in the cecal content after 3 days of treatment, which represents an at least 3-order of magnitude reduction in the microbial load. The treatment was continued for another 11 days to ensure the animals reach a PGF status. Others have reported the use of 1 to 4 antibiotics in the drinking water or by oral gavage for 3 to 21 days in mice, and they achieved reductions of 1 to 5 orders of magnitude [12-14]. The authors of these studies concluded that the treated mice resembled GF mice anatomically and physiologically [2]. The PGF rats generated herein, which experienced a similar level of reduction in the bacterial load, thus are expected to be similar to GF rats. The humanized rats produced using the procedure described above harbored gut microbiota more closely resembled to that of the human donors than the naïve rats. A similar observation has been reported by others in mice, when a highly effective antibiotic treatment was employed [9] or when using GF animals [12]. However, a smaller reduction in microbial load produced a less diverse microbiota and greater similarity to the naïve microbiota than to that of the human donors [13]. Our procedure for creating PGF rats allowed the humanderived microbiota to successfully replace the native microbiota in the rat intestinal tract (see Figure 4). Although the gut microbiota of the humanized rats is similar to that of the human donors, the native rat microbiota is robust, as indicated by the partial recovery of the native rat microbiota in the PGF rats that did not receive fecal transplant (Antibiotic/LB in Figures 5b-d).The failure of PGF rats to become humanized through coprophagia alone also confirmed this notion.
The longest stability reported in humanized rodent models of gut microbiota to date was 12 weeks [9], but the mice in that study were each associated with a single donor and had distinct microbiota and metabolomic profiles. In this study, humanized rats generated from multiple donors with diverse microbiota were depersonalized to produce a high degree of homogeneity between the humanized rats, thereby eliminating donor-specific attributes in their gut microbiota. This homogenous profile of human microbiota was stable for 15 weeks. In addition, the humanderived microbiota of the depersonalized communities appeared to be more stable than that of individual humanized lines generated from the same human donors. One potential explanation is that the human-derived microbiota created in the community setting were better able to compete against the native rat microbiota than those in the individual lines. Alternatively, the communities may have benefited from coprophagia, which essentially re-inoculated each animal with the human-derived microbiota when they consumed feces contained within the redistributed bedding.
This study also showed that mixing the donor material prior to transplantation might hinder the humanization process. The individual humanized line generated from the mixture of 3 human donors (mix-associated) were more similar to the native rat microbiota than the human microbiota after 5 weeks (Figure 5b). In addition, some members of Community 1 were mix-associated rats, and that community took longer to become homogenous and was more similar to the native rat microbiota than Community 2, which did not contain mix-associated rats (Figures 5b & 5c). Thus, the inoculum made by mixing human donor materials appears to compete poorly against the re-emerging, native rat microbiota compared to the inoculum made from a single human donor. In conclusion, we presented a procedure for generating PGF rats from commercially purchased animals, and the subsequent creation of communities of rats with a homogenous profile of humanderived microbiota that is stable for at least 15 weeks. With certain modifications, this procedure will likely be applicable for any natural or genetically modified strains of laboratory rodents, which in turn will greatly expand the models available for studying gut microbiota and its interaction with host functions.
Acknowledgment
The authors appreciated technical assistance of Chelsey Webb, Timothy Bausman, Jo Griffin, Heather Procopio, Curtis Schimmel, Katherine Ingram, Eric Perez, Amanda Short, Kirt Henderson, Amy Walters, Steve Sayson, Emily Hill, and Leah Shurte. This research was supported in part by an appointment to the Postgraduate Research Participation Program at the U.S. Air Force Research Laboratory, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and USAFRL. This report was cleared for public release on March 1, 2019 (Case Number 88ABW-2019-.612).
Funding
This study was supported by the Office of Naval Research, Award # FA86501026A02.
Declaration of Conflicting Interest
The Authors report that no conflict of interest is involved in this research or its publication.
Supplemental Data
Bioassays for Selected Markers of Host Response
Several markers of the host functions and host-microbiota interaction in the humanized rats (including intestinal myeloper oxidase (MPO) activity, serum level of lipopolysaccharide binding protein (LBP), proinflammatory cytokines and hypothalamus-pituitary- adrenal (HPA) axis hormones) were examined.
Methods
Serum and tissue samples were collected post-mortem at the
timepoints indicated in “Materials and Methods” and stored at -80
°C until use. The small intestine homogenate collected as described
under “Determination of Bacterial Titer” was assayed for MPO
activity using the Myeloperoxidase Activity Assay Kit (Abcam,
Cambridge, MA), according to the manufacturer’s instructions.
The following bioassays were performed using serum samples
following the manufacturers’ instructions:
a) LPB (Lipopolysaccharide Binding Protein Kit from
MyBioSource, San Diego, CA);
b) Adrenocorticotropic hormone (ACTH ELISA Kit from
MyBioSource);
c) Corticosterone (Corticosterone ELISA Kit from Arbor
Assays, Ann Arbor, MI);
d) Corticotrophin releasing factor (Corticotrophin Releasing
Factor ELISA Kit from Kamiya Biomedical, Seattle WA); and
e) A panel of proinflammatory cytokines (VPLEX Plus Proinflammatory
Panel 2 Rat Kit from Meso Scale Diagnostics, Rockville,
MD).
Statistical analysis was accomplished using Prism 7 for
Windows, version 7.05 (GraphPad Software, La Jolla, CA). One-way
ANOVA with Tukey multiple comparison was used with alpha of
0.05.
Result
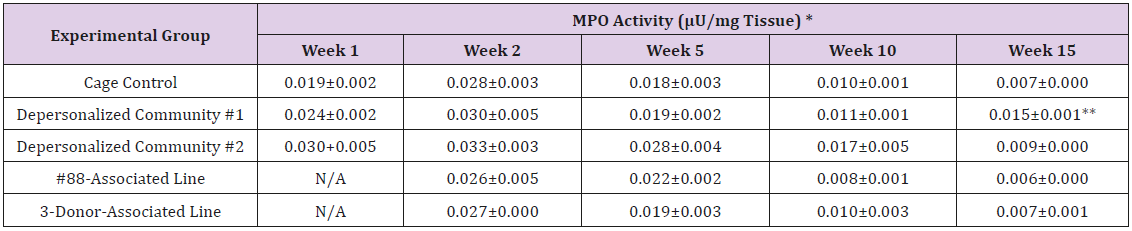
Supplemental Table 1: Myeloperoxidase (MPO) Activity in Small Intestine Tissue Homogenate.
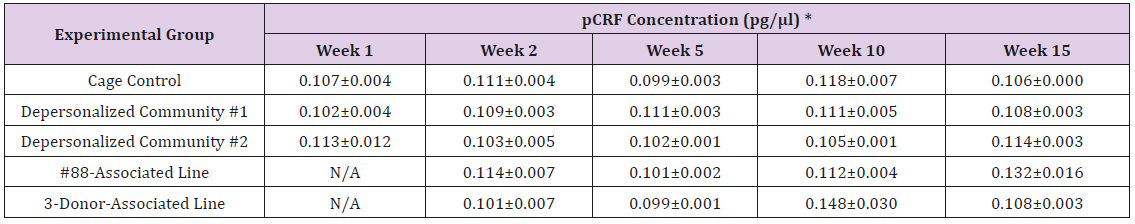
Note: *Mean ± SE; 1 U of MPO activity = Amount of MPO needed to convert 1.0 μ mol of substrate to product per minute at 25°C; ** Statistically significant different from cage control (p < 0.05).
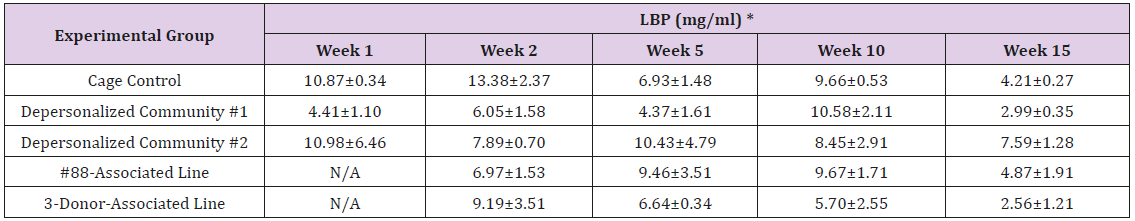
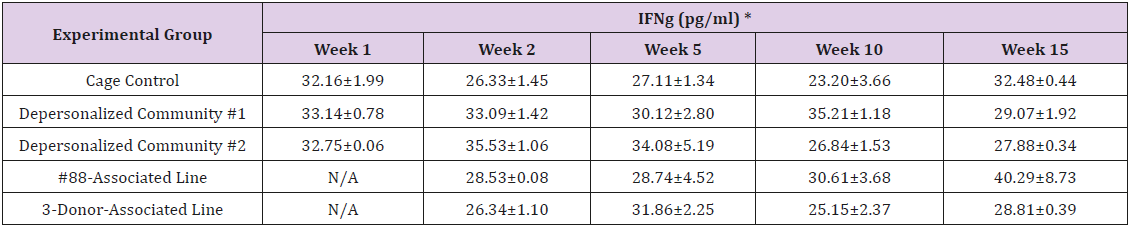
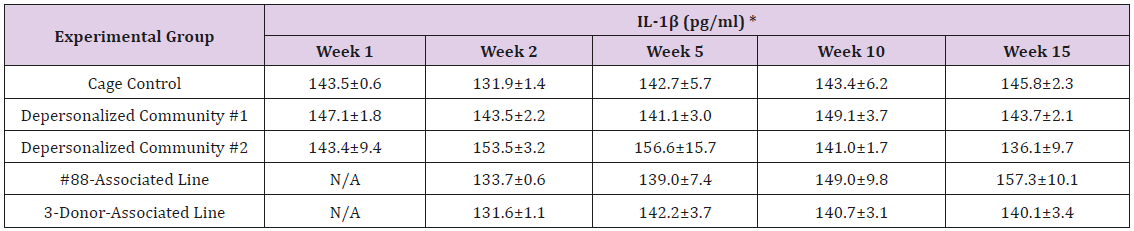
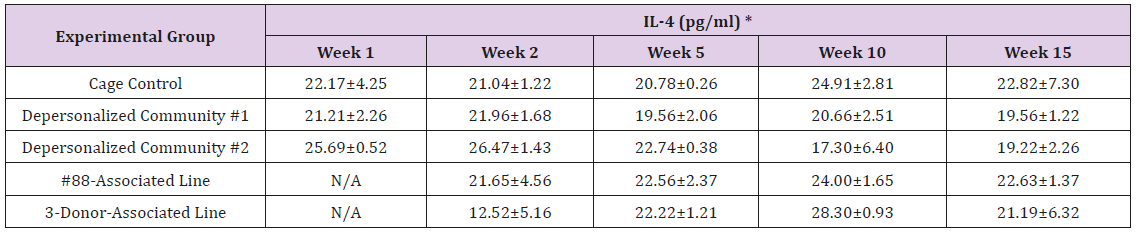
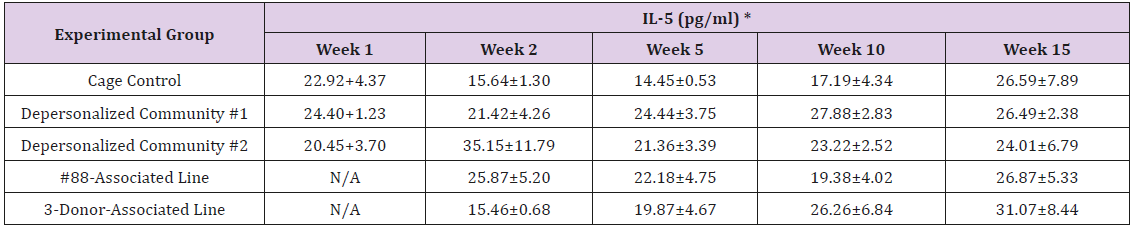
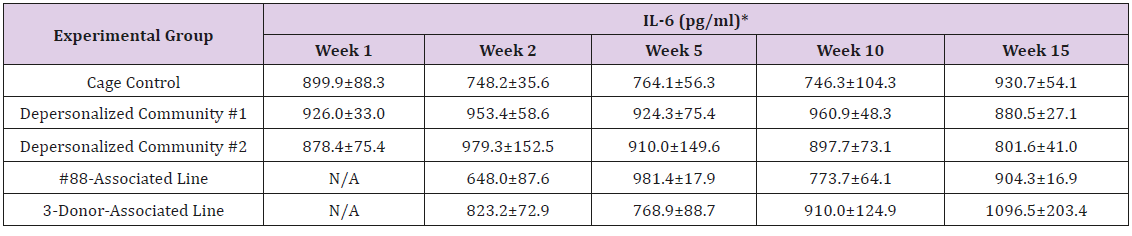
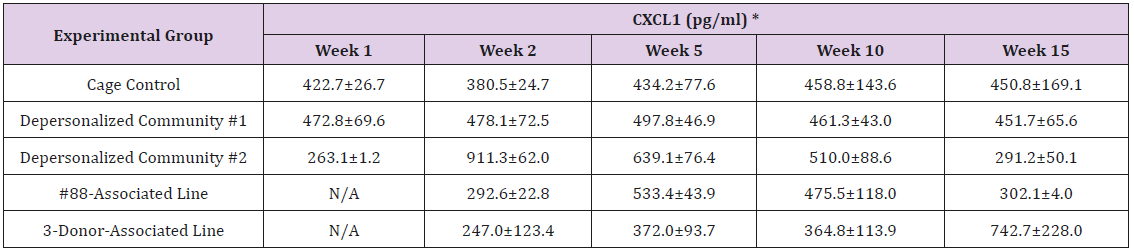
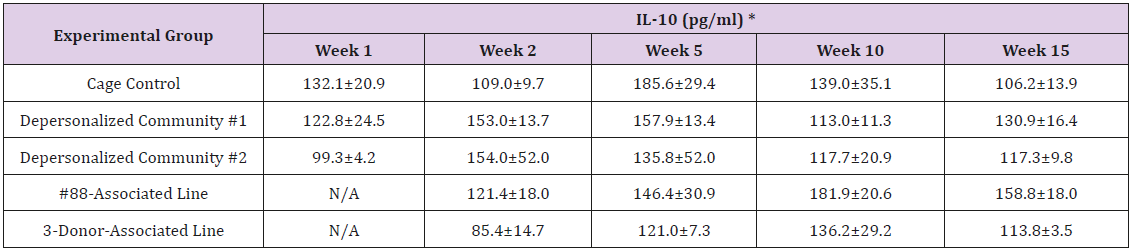
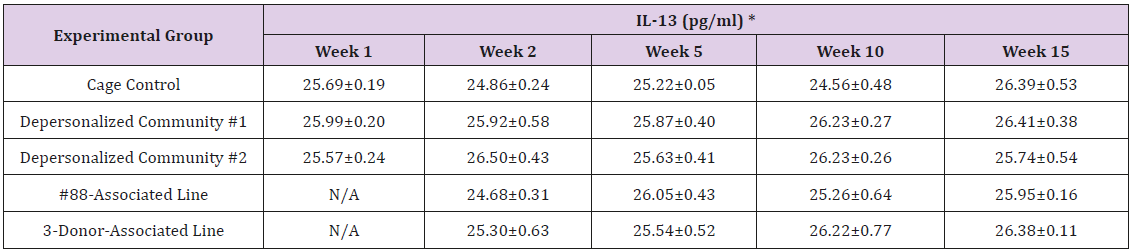
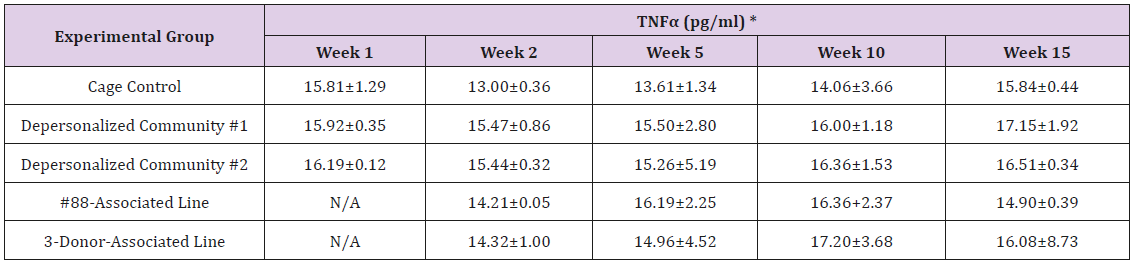
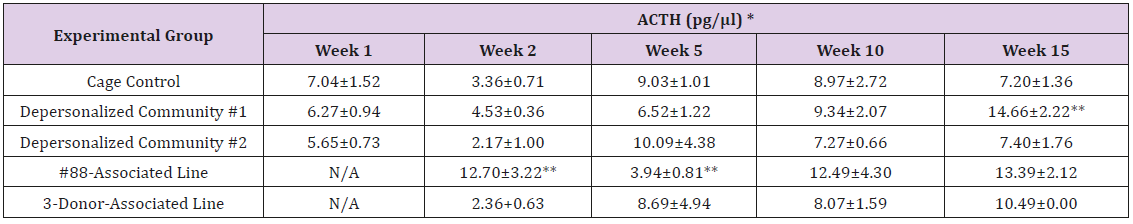
The results of these bioassays showed that there were no biologically relevant changes that would indicate a negative reaction to the humanization process or a significant change in host function or interaction with the gut microbiota. MPO activity in the intestinal tissues was relatively unchanged except for community #1 at 15 weeks, which had an increase in activity that might indicated a reaction to bacteria in the tissue (Supplemental Data Table 1). There was no change in the serum level of LBP, suggesting that the barrier function of the gastrointestinal tract remained relatively intact during the 15 weeks of depersonalization (Supplemental Data Table 2). This was confirmed by culture data that did not find any bacterial cells in any of the extra-intestinal tissues including mesenteric layer, mesenteric lymph nodes, spleen and liver (data not shown). There were some small changes in the cytokine levels, but none was statistically significant after the correction for multiple testing (Supplemental Data Table 3). For the hormones of the HPA axis, only sporadic changes in the level of adrenocorticotropic hormone (ACTH) and corticosterone occurred and were not likely a specific stress response to the experimental procedure or the colonization of the human microbiota (Supplemental Data Table 4). In the #88-Associated Line, ACTH was significantly higher at 2 weeks and lower at 5 weeks than the cage control; and corticosterone levels were significantly higher than control at 10 weeks. The Depersonalized Community 1 had a significantly higher level of ACTH than the cage control at 15 weeks. The 3-donor mixassociated line had significantly higher corticosterone than the control at 15 weeks. Since these changes did not follow a specific pattern and did not correlate with other markers, the biological significance of these changes is not completely understood at this point.Taken together, these results suggested that the human microbiota probably interacts with the rat intestinal tract in a similar way as the native rat microbiota and is unlikely to cause any significant change in the host functions.
Supplemental Table 3: Serum Levels of Pro- and Anti-Inflammatory Cytokines.
Supplemental Table 4: Serum Levels of Hypothalamic–Pituitary–Adrenal Axis Hormones:
Supplemental Table 4b: Adrenocorticotropic Hormone.
Note: *Value = Mean ± SE; **difference from cage control is statistically significant (p<0.05).
Supplemental Table 4c: Corticosterone.
Note: *Value = Mean ± SE; **difference from cage control is statistically significant (p<0.05).
References
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. (2005) Diversity of the human intestinal microbial flora. Science 308: 1635-1638.
- Rakoff Nahoum S, Paglino J, Eslami Varzaneh F, Edberg S, Medzhitov R (2004) Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118: 229-241.
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, et al. (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101: 15718-15723.
- Cryan JF, Dinan TG (2012) Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 13: 701-712.
- Collins J, Auchtung JM, Schaefer L, Eaton KA, Britton RA (2015) Humanized microbiota mice as a model of recurrent Clostridium difficile disease. Microbiome 3: 35-015-0097-2.
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, et al. (2009) The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1: 6ra14.
- Becker N, Kunath J, Loh G, Blaut M (2011) Human intestinal microbiota: characterization of a simplified and stable gnotobiotic rat model. Gut Microbes 2: 25-33.
- Mooser C, Gomez de Aguero M, Ganal Vonarburg SC (2018) Standardization in host-microbiota interaction studies: challenges, gnotobiology as a tool, and perspective. Curr Opin Microbiol 44: 50-60.
- Hintze KJ, Cox JE, Rompato G, Benninghoff AD, Ward RE, et al. (2014) Broad scope method for creating humanized animal models for animal health and disease research through antibiotic treatment and human fecal transfer. Gut Microbes 5: 183-191.
- Ji SK, Yan H, Jiang T, Guo CY, Liu JJ, et al. (2017) Preparing the Gut with Antibiotics Enhances Gut Microbiota Reprogramming Efficiency by Promoting Xenomicrobiota Colonization. Front Microbiol 8: 1208.
- Staley C, Kaiser T, Beura LK, Hamilton MJ, Weingarden AR, et al. (2017) Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome 5: 87-017-0306-2.
- Wos Oxley M, Bleich A, Oxley AP, Kahl S, Janus LM, et al. (2012) Comparative evaluation of establishing a human gut microbial community within rodent models. Gut Microbes 3: 234-249.
- Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, et al. (2010) Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res 20: 1411-1419.
- Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, et al. (2011) Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One 6: e17996.
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335-336.
- De Santis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, et al. (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069-5072.
- Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228-8235.
- Vázquez Baeza Y, Pirrung M, Gonzalez A, Knight R (2013) EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2: 16-217X-2-16.
- (2012) Human Microbiome Project Consortium (248 Collaborators). Structure, function and diversity of the healthy human microbiome. Nature 486: 207-214.
- Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R (2011) UniFrac: an effective distance metric for microbial community comparison. ISME J 5: 169-172.
- Afra K, Laupland K, Leal J, Lloyd T, Gregson D. (2013) Incidence, risk factors, and outcomes of Fusobacterium species bacteremia. BMC Infect Dis 13: 264-2334-13-264.

 Research Article
Research Article