Abstract
Background: Bladder cancer is one of the dominating causes of death worldwide. The identification of microRNA functions in carcinogenesis has given a new direction for the development of novel biomarkers.
Aims: We have investigated seven microRNAs (miR-124, miR-126, miR-139, miR- 145-3p, miR-145-5p, miR-152, miR-182) in order to define their clinical relevance in diagnosis and prognosis of bladder cancer patients. We have analyzed expression levels of these microRNAs in tumor samples versus normal tissues, as well as correlation between their expression levels and staging and lymph node metastasis.
Methods: Expression levels of selected microRNAs were assessed by TaqMan RTPCR assay. The data obtained were revealed using GraphPad Prism 6.01 software which enabled the statistical analysis of the correlation between the miRNA expressions and tumor stage.
Results: We have found a consistent epigenetic signature in tumor samples including significant downregulation of miR-126, miR-139, miR-145-5p, miR-145-3p, miR-152, as well as upregulation of miR-182 compared to normal tissues. The expressions of miR- 126, miR-139 and miR-152 were significantly downregulated in patients with T3N0 stages compared to patients with T3N positive stages. Our preliminary data show that selected miRNAs are clinically relevant for monitoring the urinary bladder cancer patients.
Keywords: MicroRNAs; Epigenetics; Urinary Bladder Cancer (UBC); Cancer Stage
Abbreviations: TSG: tumor suppressor genes; OG: oncogenes; TS-miRNA: tumor suppressive microRNA; Onco-miRNA: oncogenic microRNA; TNM: Tumor Node Metastasis; UBC: urinary bladder cancer
Introduction
Bladder cancer is an important cause of death worldwide. This type of cancer is a major concern related to life expectancy worldwide, also in Romania. According to WHO publication, in 2017 a number of 1651 of bladder cancer deaths have been reported in Romania, representing 0.70% of total urinary bladder cancer deaths [1]. Presently, a lot of research data are published reporting insight aspects of tumorigenesis in bladder cancer, which are used in establishing diagnosis and prognosis of this devastating disease. The most of these reports focus mainly on non-genetic or epigenetic factors linked with prevention and personalized targeted treatments [2,3]. Also, much effort is involved in delimiting the stages and grades of tumorigenesis in bladder cancer, that are already classified by WHO 1973 and further completed in 2016, based on standardized pathological criteria [4]. The biological mechanism of bladder cancer tumorigenesis in terms of epigenetic factors involvement. It is already accepted by the worldwide cancer epigenetic research that tumorigenesis is a process implying both genetic and epigenetic changes. Malignant tumors have molecular features which have developed in successive steps resulting the excessive growth of cell clones in a normal tissue. The main feature of cancer cells is to be dependent on the abnormal pattern of gene expression, involving two essential types of genes: tumor suppressor genes (TSG) and oncogenes (OG).
The reprogramming of their expression is realized through their epigenetic state: while TSG are active in any normal cell, they may be aberrantly silenced by either DNA methylation and histone covalent modifications of their promoter chromatin or microRNA high level expression in cytoplasm. By contrast, proto-oncogenes are normally silenced and aberrantly activated as oncogenes in tumor cells. Based on these processes, the investigation of microRNAs involvement in the regulation of several genes during cancer initiation and progression has led to the assumption of the microRNA’s role in cancer as TSG or OG [5]. Therefore, Lu et al. have proposed a molecular classification of tumor according to changes in microRNA expressions [6]. Differential expression of microRNA genes between malignant and normal cells is a complex process which underlies a lot of factors such as microRNA control of oncogenes and tumor suppressor genes and genomic location of microRNAs in cancer-associated regions. TS-miRNAs have antiproliferative roles by their ability to negatively regulate messenger RNA (mRNA) of oncogenes through their expression level as compared with the normal tissue. This role is proved by their under-expression which is correlated with overexpression of the oncogenes controlled by the TS-miRNAs.
This process is in accordance with tumor development [7]. Particular epigenetic regulation of mRNAs corresponding to TSG or OG may characterize differently miRNAs as tumorigenic (having OG role), or tumor suppressive (having TSG role). Post transcriptional epigenetic control of OG or TSG mRNA translation may be positive in the context of miRNA under-expression and negative in the context of miRNA overexpression. Therefore, a miRNA species that negatively regulate an OG mRNA became a TS-miRNA with tumor suppressive role. By contrast, its under-expression is able to positively regulate an OG mRNA translation defining its OG or tumorigenic role. In the same way the overexpression of miRNA species that negatively regulating a TSG mRNA has tumorigenic or OG role, while its under-expression towards the same TSG mRNA may turn in to opposite tumor suppressive or TSG role.7 In some cases, the same microRNA may have dual activities acting either as an OG or TSG in a tissue-specific manner, in different stages of the tumor progression [8]. Previously, we have reported our results regarding the expression profile of certain microRNAs (miR-145- 3p, miR-145-5p, miR-152 and miR-182) in Romanian patients with urinary bladder cancer [9].
This work uses these preliminary results and investigates the clinical relevance of additional miRNAs species (miR-124, miR- 126 and miR-139) in our patients. The present approach aims to figure the possibility of defining an epigenetic molecular diagnosis algorithm. Moreover, the new results establish the tumorigenic role (as tumor suppressor gene or oncogene) for each microRNA. Detailed discussions of these aspects compared with statistical analysis values are reported as the final criteria for selection of relevant microRNAs. The extended microRNAs expression markers could provide more relevant information. We are interested to seek for the relevance of the selected miRNAs in delimitating the stages during bladder cancer progression related to TNM (Tumor, Node, Metastasis) classification. Modulation of microRNA expression is increasingly considered to be an important mechanism by which tumor suppressor proteins and oncoproteins exert some of their effects with important results regarding the stages of the carcinogenesis. This work considered such concepts by a particular statistical tool that enabled a comparison between the expressions of miRNA species during the various carcinogenesis stages.
Patients and Methods
Patients
In this study we have included ninety patients with urinary bladder tumors (73 males and 17 women) who were admitted in Fundeni Clinical Institute, Centre of Urology, Dialysis and Kidney Transplantation from May 2017 to November 2018. The median age was 66 years (ranging from 36 to 82). The diagnosis was based on clinical, laboratory and imagistic examinations. All patients had undergone cystectomy and during the surgery were collected ninety bladder tumor tissues and twenty-two adjacent normal tissues. Collected tissue samples were immediately stored at -80°C. Also, all samples were processed for pathological examination by standard diagnostic procedure, according to TNM classification. Informed written consent was obtained from all patients, and the research protocol was approved by the Fundeni Clinical Institute Ethical Committee.
Methods
RNA Extraction and Amplification: Total RNA was isolated from frozen tumor and normal tissue samples using a mirVana miRNA Isolation Kit (Invitrogen) according to manufacturer’s instructions and the concentration was assessed by a Qubit Fluorometer (Invitrogen). Total RNA samples were reverse transcribed using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems USA). Quantitative real-time PCR for selected mature microRNAs was performed according to the manufacturer’s instructions for TaqMan MicroRNA Assays by using TaqMan Universal Master MixII, no. UNG (Applied Biosystems) on Applied Biosystems 7300 Real Time PCR system. The expression level values were normalized to the endogen control expression of RNU48. All reactions were done in duplicate.
Statistical Analysis: The levels of microRNA species in investigated patient samples were expressed relative to control samples using the double delta CT method. Statistical analysis was performed using GraphPad Prism 6.01 software. The results were expressed as mean ± SEM (Standard Error of Mean). P value was calculated using Student t test. The P-value < 0.05 was considered statistically significant.
Results
MicroRNA Expressions Profile Between Tumor Samples and Normal Tissue Samples
Investigation of urothelial tumors involved the evaluation of selected microRNAs relative expression in tumor samples compared with their expression in normal tissues. We have found that the microRNAs expression profiles differed between tumor and normal bladder tissues:
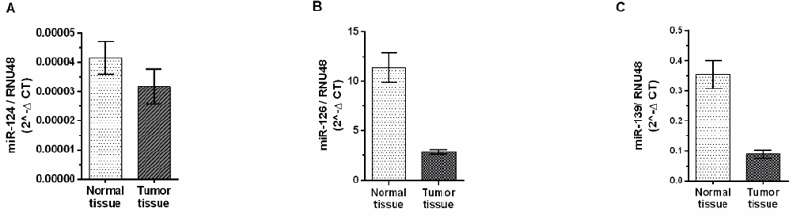
a) The expression levels of miR-126, miR-139, were significantly downregulated in tumor samples compared to normal samples (P<0.0001) (Figure 1)
b) miR-124 was also downregulated in tumor tissues although its expression was not statistically significant (P=0.2409) (Figure 1)
c) The expression levels of miR-145-3p, miR-145-5p and miR-152 remained significantly under-expressed (P<0.001 in tumor bladder tissues compared to normal bladder tissues [9]
d) miR-182 was significantly upregulated in bladder cancer tissues versus normal tissues [9].
Figure 1: Expression profile of miR-124, miR-126 and miR-139 presented as 2-ΔCT of the miRNA expression level in tumor and matched adjacent normal bladder tissues.
MicroRNA Expression Analysis According to Cancer Stage
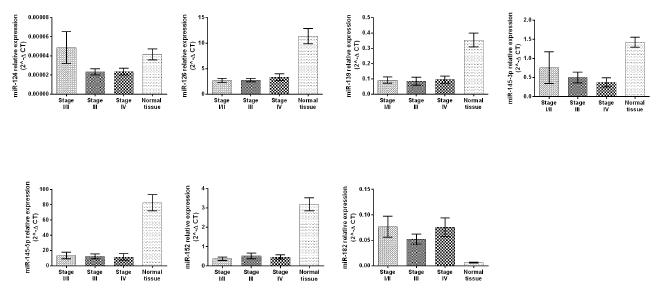
The patients were divided by their cancer stage as follows: I, II, III and IV (represented by 9, 21, 43 and 17 patients), respectively corresponding to the expression levels of the microRNA’s species. The expression levels have been divided in three groups comprising the following stages: stage I/II, stage III and stage IV. In this study, we have found a significant correlation of miR-126, miR-139, miR- 145-5p, miR-152 and miR-182 expression levels with tumor stages I/II, III and IV, compared with normal tissues (P<0.0001) (Figure 2). Also, for miR-145-3p and miR-124 we obtained significant correlations of change in expression only in stages III and IV compared with normal tissues (P<0.0001) (Figure 2). We have observed a tendency to a decreased miR-124 expression level in tumor stages III and IV compared with normal bladder tissue. (P=0.096 respectively, P=0.0117) (Figure 2).
Figure 2: Correlation between the miR-124, miR-126, miR-139, miR-145-p, miR-145-5p, miR-152, miR-182 expression levels and tumor stages presented as 2-ΔCT. Error bars represent SEM.
MicroRNA Expression Analysis According to Every Stage of Primary Tumor Development (T)
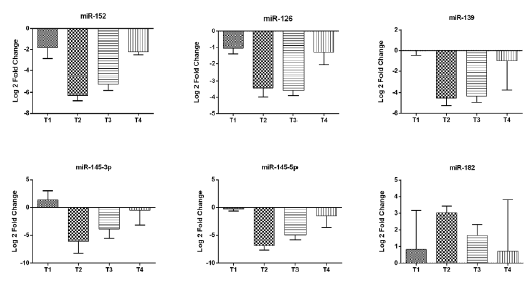
Further, the statistical analysis was approached considering the samples divided in other four groups T1, T2, T3 and T4 (according to T from TNM classification). Only tissues from patients without metastasis were analyzed. In this analysis we did not obtained statistically significant results. It was observed a tendency for decreased expression level in T2 group compared with T1 group for miR-126 (P=0.0612), miR-139 (P=0.0830), miR145-5p (P=0.0767). Also, miR expression levels were lower in T3 group compared with T1 group for miR-126 (P=0.0601), miR-139 (P=0.0845), miR145- 5p (P=0.0845). miR-152 level presented a tendency to decreased expression level in T3 group compared with T4 (P=0.0869). miR- 182 expression was a slightly increased in T2 patients than T3 ones (P=0,0931). The results are presented in (Figure 3).
Figure 3: Analysis of microRNAs expression level according to T stage in bladder cancer tissues: miR-152, miR-126, miR-139, miR-145-3p, miR-145-5p, miR-182 presented as Log 2-Fold Change. Error bars represent SEM.
MicroRNA Expression Analysis According to Lymph Node Metastases
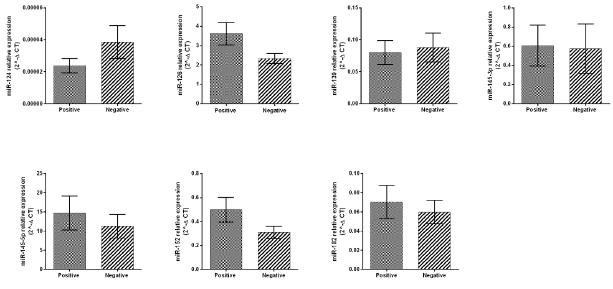
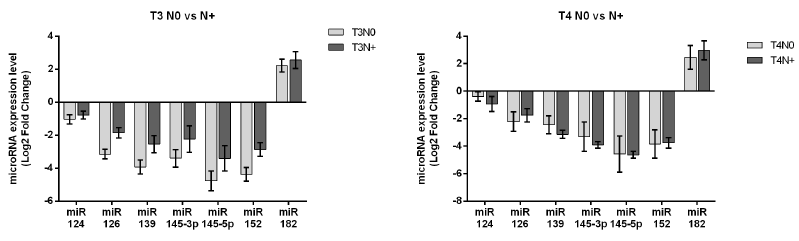
This analysis involved the comparison of microRNAs expression in tumor tissues with different, invasive/noninvasive. In total, 40 (44.5%) patients had positive and 50 (55.5%) had negative lymph node metastases. With the exception of miR-126, for which we obtained a borderline result (P=0.0512), not significantly correlation between positive or negative lymph nodes and microRNA expression levels were observed (Figure 4). In addition, analyzed tissues were divided according to tumor development stage as T2, T3 and T4 and subdivided according to the presence (N+)/absence (N-) of lymph node metastasis. Further, the correlation between microRNA levels and lymph node metastases were evaluated in tumor development stages T3 and T4. The results showed the statistically significant decrease of miR-126 (P=0.0233), miR-139 (P=0.0416) and miR-152 (P=0.0273) in T3N0 patients compared with T3N+ subjects (Figure 4). For miR-145-5p we obtained a borderline result (P=0.0514) (Figure 5). Within the T4N0 and T4N+ groups not significantly difference in the expressions of the analyzed microRNA species were observed (Figure 5).
Figure 4: Relative expression of miR-124, miR-126, miR-139, miR-145-3p, miR-145-5p, miR-152, miR-182 between positive and negative lymph node metastases tissues, presented as 2-ΔCT.
Figure 5: Analysis of miR expression level according to lymph node metastases versus metastasis-free patients in bladder cancer tissues: T3N0 versus T3N+ and T4N0 versus T4N+.
Patients monitoring after cystectomy by imagistic examination lead to their classification in two categories: patients with metastases (39%) and patients without metastases (61%). microRNA expression levels comparison between the two groups highlighted a significant increasing of miR-182 (P=0.0039) at patients with metastases compared with patients without metastases. In contrast, miR-126 showed a significant decrease in patients without metastases (P=0.0081). For the other microRNA species no significant differences were obtained.
Discussion
Recent studies in the field of epigenetics have contributed to a better understanding of carcinogenesis processes and therefore to a better characterization of urinary bladder tumors. The newly discovered molecular features of tumors could be used in defining diseases prognosis and in anticancer therapeutic schemes. Posttranscriptional control of tumor suppressor genes and oncogenes is performed in cytoplasm by the activity of OG-miRNAs and TSGmiRNA species. Consequently, expression level of microRNAs involved in such processes could be considered a differentiation marker between normal and tumor tissues. Interpretation of our results in this context is based on data from literature [10] which explain the oncogenic/tumor suppressor roles of microRNAs according to expression level. Thus, microRNA overexpression in cancer could indicate its oncogenic role, promoting cancer development by a negative control of tumor suppressor genes and genes that control cellular differentiation and apoptosis. Underexpression of a microRNA species in cancer could indicate its acting like tumor suppressor genes and inhibition of cancer by controlling oncogenes and genes that control cellular differentiation and apoptosis.
The evaluation of selected miRNA expression levels between tumor and normal bladder tissues led to their classification in two subgroups which define the molecular mechanism underlying their tumorigenic function: subgroup marked by under-expression and the subgroup marked by overexpression. The common element of these microRNA species in terms of mode of action is the negative control of the expression (mRNA translation) of oncogenic or tumor suppressor proteins. Subgroup defined by under-expression was represented by the most microRNA species: miR-124, miR- 126, miR-139, miR-145-3p, miR-145-5p and miR-152. Their role has been suggested to be the promotion of oncogenes expression and they are free to perform their tumorigenic function while the cancer progression occurs. miR-182 is the only representative of subgroup defined by overexpression, its oncogenic role may be based on an inhibitory mechanism of the tumor suppressor genes expression. A correlation of miR-124 under-expression tendency was observed with advanced tumor stages, which could suggest that this is contributing to tumor aggression by loss of control of targeted oncogenes. Such results are confirmed by numerous scientific reports. Neuronal-enriched miR-124 have a role in modulation of innate and adaptive immune systems and in inflammatory diseases [11,12]. Its downregulation has tumorigenic effects in multiple cancer types: oral carcinoma, nasopharyngeal carcinoma, hepatocellular carcinoma, breast cancer, lung cancer, bladder cancer [13-19]. Numerous studies conducted in bladder tumor tissues and in bladder tumor cell lines have revealed the tumor suppressor function of miR-124 by its ability to target mRNA of genes involved in complex biological processes underlying cell transformation.
The identification of miR-124 tumor suppressor role by its direct binding to the 3’-UTR region of mRNA of the ROCK1, CDK4, AURKA, UHRF1, CAV1 genes has established the relationship between it and bladder cancer. Thus, miR-124 under-expression lead to the increase of ROCK1, AURKA, UHRF1, CAV1 expression levels that are correlated with the progression of bladder cancer by proliferation, apoptosis suppression and metastasis [20-23]. In relation with TNM stages, the tumor suppressor miR-126 was significantly under-expressed in all stages of bladder tumors tissues compared with normal bladder tissues analyzed. Statistical analysis of the miR-126 level revealed its tendency to be downregulated depending on tumor progression: miR-126 expression was lower in T2 and T3 groups compared with T1 group. In the case of lymph node metastases, we obtained a borderline significance for miR-126 increased expression in group of tumors that have spread to the lymphatic flow compared with metastases-free tumors. miR- 126 expression level was significantly lower in T3N0 compared with T3N+. Considering the initial hypothesis according to that the decreasing of tumor-suppressor miRNA expressions are leading to increased aggressiveness, this is not confirmed by the lower miR- 126 expression levels in tumors without lymphatic invasion than in lymphatic invasion tumors. An explanation for this could be related to the tumor tissue heterogeneity regarding the stages of neoplastic progression, which is also based on pathological staging. In this way, the population of cells in which the microRNA is expressed at a certain value compared to normal cells cannot be controlled by such a statistical approach.
However, another biological explanation would refer to the intense dynamics of reprogramming processes of gene function in neoplastic progression. These would involve the activation of other oncogenes and the inactivation of other suppressor tumor genes, against which the studied microRNA species did not have control. Downregulation of miR-126 expression was also observed in various solid cancers (colorectal, pancreatic, thyroid, breast, esophageal, kidney and bladder cells), but not in leukemia where the expression of miR-126 varied depending on the leukemia subtype [24]. Identification of numerous targets for miR-126 in different tumors highlights its major impact in cancer development and progression. In most cases, miR-126 acts as tumor suppressor [25]. However, miR-126 have a complex behavior acting either as oncogene, or as tumor suppressor in different physiological and pathological states [26]. The tumor suppressor role of miR- 126 in the progression of bladder cancer was first described by the identification of inverse correlation between its expression and metalloproteinase ADAM9 expression. Increasing of ADAM9 level expression due to miR-126 under-expression is involved in increasing invasive potential of bladder cancer cells [27].
miR-126 has important regulatory function in PI3K/Akt pathway transduction, which is thought to play a major role in bladder carcinogenesis [28]. Inhibition of PIK3R2 expression and the PI3K /Akt signaling pathway by miR126 suggests that it can negatively regulate targeted gene and inhibit the signaling pathway PIK3R2/PI3K/Akt [29]. miR-139 is located on chromosome 11q13.4 and has anti-oncogenic and anti-metastatic activity. Deregulation of miR-139 expression was reported in many diseases, including cancer.30, 31 Under-expression of miR-139 was first shown in the process of neurodegeneration and later, its role in cancer diagnosis, prognosis and therapy was identified.31 miR-139 downregulation in cancer was significantly associated with unfavorable prognosis and with metastatic tumors features including venous invasion, absence of tumor encapsulation and poor differentiation [30]. Cao discovered that p53 regulates miR-139 transcription which suppress the expression of PDE4D oncogene and, also induce cell growth arrest mediated by cAMP/BIM. They suggested that deregulation of p53-miR-139-PDE4D-cAMP-BIM pathway could be associated with tumorigenesis [31,32]. Other studies confirmed our findings regarding the participation of miR-139 in the migration and invasion of bladder cancer cells.
Yonemori et al. have identified that MMP11 is directly targeted by miR-139. Under-expression of miR-139 has been correlated with MMP11 overexpression which is involved in regulation of many oncogenes related to migration and invasiveness of bladder cancer cells [33]. Luo have demonstrated that miR-139 inhibits bladder tumor growth by directly blocking of Bmil expression. miR- 139 under-expression leads to the activation of Bmi1 oncogenic role in the bladder cancer tumorigenesis, which is associated with tumor growth stimulation and with the maintaining cancer stem cells proprieties [34]. Our study also showed that the expression level of miR-139 was significantly downregulated in all bladder tumor stages compared with normal bladder tissues. In addition, a significant result was obtained for expression level of miR-139 between T3N0 versus T3N positive. Therefore, the expression level was significantly lower in T3N0 compared with T3N positive. Under-expression of miR-139 in tumors that have not spread in lymph nodes compared with the tumors that have spread to lymph nodes, could suggest its intensive tumorigenic role in tumor progression.
Thus, a decrease of miR-139 level could lead to loss of control of some oncogenes and consequently, to activation of novel molecular signaling pathways where miR-139 function is less involved. Based on correlative analyzes of miRNA expression, we suggest that miR-139 could serve as a progression marker for bladder tumors and also could represent a negative prognostic biomarker. Additionally, the role of miR-139 in tumor progression can also be suggested by comparative analysis of its expression between groups T1 vs T2 / T3. The miR-139 level has shown a tendency to decrease in T2 and T3 compared to T1. The assessment of the tumorigenic role of the microRNA species approached in the initial phase of this study (miR-145-3p, miR-145-5p, miR-152, miR-182), from the point of view of their correlation with tumor stages and lymphatic invasion provided new data on their dynamics in tumor progression. Our study has shown that the expression of miR-145- 3p was significantly decreased in tumor stages III and IV compared with normal bladder tissues, suggesting that it has an intensive tumorigenic role in advanced tumors. miR-145-3p tends to be downregulated in T3N0 compared with T3N+.
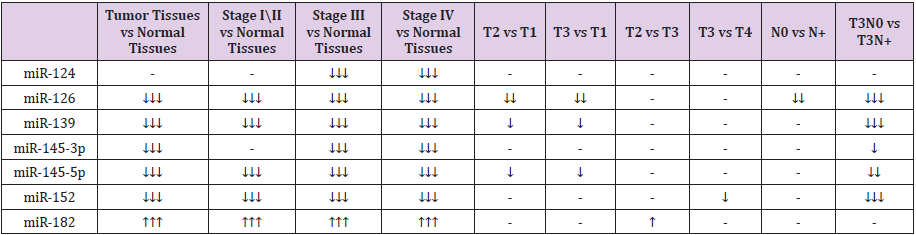
The expression level of miR-145-5p was significantly lower in all tumor stages, suggesting that its tumor suppressive function is altered since the first tumor stage. In relation to the extent of primary tumor (T), it was observed a tendency to decrease of miR- 145-5p expression in groups T2 and T3 compared with T1 group. In addition, borderline result was obtained for miR-145-5p expression in the case of metastasis to the lymph nodes. Its expression level was decreased in T3N0 patients compared with T3 metastasesfree subjects. miR-152 expression was downregulated in group of bladder tissues versus normal tissues. In accordance with this result, we found that miR-152 was strongly downregulated in all bladder cancer stages. In the case of primary tumor extension (T), it was observed its tendency to be more downregulated in T3 group than T4. The negative correlation of miR-152 with T stage could suggest its tumorigenic behavior, dependent on its expression; it decreases with tumor growth. By the other hand, in the correlative analysis with lymph node metastases, we obtained a significant decrease of miR-152 expression in T3N0 compared with T3N+. miR-182 expression was changed in all tumor stages compared with normal tissues and presented a tendency to be overexpressed in T2 group compared with T3. Finally, only several miRNAs proved statistical significance in their association with bladder cancer tumor progression. These results are summarized in the diagram of tumor progression in Table 1.
Table 1: The most relevant variations of selected microRNA expressions. ↓↓↓ = significant (P <0.005), ↓↓ = borderline (0.05), ↓ = tendency (>0.07<0.1).
Conclusions
Deregulation of molecular mechanisms control can be caused by synergic cooperation of several microRNA species which modulate the mRNAs transcripts involved in key cellular processes. Changes of miRNA expressions may be drivers of carcinogenesis processes but may also be the result of tumor progression. In this way, the expression profiles of the microRNA species could become useful biomarkers in cancer patient’s diagnosis. In addition, such results may be useful in personalized therapy decisions and also in cancer prevention by direct use of microRNA species in diagnosis. In this study, the most representative microRNA in terms of prognostic prediction is represented by miR-182. Increasing of miR-182 expression level to patients with metastases suggest its role in tumor progression and metastasis and, also could be correlated with an unfavorable prognostic. The selection and inclusion of microRNA species on a panel representative for each neoplastic stage impose an extended testing on a larger group of patients.
Our results are important for the elaboration of personalized diagnosis scheme of bladder cancer tumors which include staging of the cancer progression in both terms of TNM criteria and molecular miRNA expression profiles. Post-transcriptional modification control in different stages of neoplastic progression is highlighted by dynamic of microRNAs expression. Evaluation of microRNAs expression and their role in tumorigenesis is a clinically relevant marker which could correlated with tumor development for a better approach of bladder cancer management. Molecular characterization of urinary bladder tumors by the selected microRNAs panel investigation provide new perspectives in precision medicine by improving the diagnostic and prognostic prediction. Also, they may be efficiently used for the innovative molecular treatment of tumor tissues by injecting precise synthetic oligonucleotides able to modify the in vivo miRNA expression that could have a particular tumorigenic activity.
Declaration of Conflicting Interests
The authors declare that there are no conflicts of interest.
References
- https://www.worldlifeexpectancy.com/romania-bladder-cancer.
- Casadevall D, Kilian AY and Bellmunt J (2017) The prognostic role of epigenetic dysregulation in bladder cancer: A systematic review. Cancer Treatment Reviews 61: 82-93.
- Giannopoulou AF, Velentzas AD, Konstantakou EG (2019) Revisiting Histone Deacetylases in Human Tumorigenesis: The Paradigm of Urothelial Bladder Cancer. Int J Mol Sci 20(6).
- Compérat EM, Burger M, Gontero P (2018) Grading of Urothelial Carcinoma and The New “World Health Organisation Classification of Tumours of the Urinary System and Male Genital Organs 2016”. Eur Urol Focus 5(3):457-466.
- Shah MY, Ferrajoli A, Sood AK (2016) microRNA Therapeutics in Cancer - An Emerging Concept. EBio Medicine 12: 34-42.
- Lu J, Getz G, Miska EA (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043): 834-838.
- Seven M, Karatas OF, Duz MB (2014) The role of miRNAs in cancer: from pathogenesis to therapeutic implications. Future Oncol 10(6):1027-1048.
- Garzon R, Calin GA, Croce CM (2009) MicroRNAs in Cancer. Annual Review of Medicine. Annu Rev Med 60: 167-179.
- Iacob MM, Petcu C, Vassu Dimov T (2018) MicroRNA expression profiles in Romanian patients with urinary bladder cancer preliminary results. Immunogenetics: Open Access 3(1).
- Zhang B, Pan X, Cobb GP (2007) microRNAs as oncogenes and tumor suppressors. Developmental Biology. Dev Biol 302(1): 1-12.
- Sun Y, Luo ZM, Guo XM (2015) An updated role of microRNA-124 in central nervous system disorders: a review. Front Cell Neurosci 9: 193.
- Qin Z, Wang PY, Su DF (2016) miRNA-124 in Immune System and Immune Disorders. Front Immunol 7: 406.
- Hatziapostolou M, Polytarchou C, Aggelidou E (2011) An HNF4α-miRNA Inflammatory Feedback Circuit regulates Hepatocellular Oncogenesis. Cell 147(6): 1233-1247.
- Li X, Fan Q, Li J (2017) MiR-124 down-regulation is critical for cancer associated fibroblasts-enhanced tumor growth of oral carcinoma. Exp Cell Res 351(1): 100-108.
- Peng XH, Huang HR, Lu J (2014) MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Mol Cancer 13: 186.
- Cai QQ, Dong YW, Wang R (2017) MiR-124 inhibits the migration and invasion of human hepatocellular carcinoma cells by suppressing integrin αV expression. Scientific Reports 7 :40733.
- Liang YJ, Wang QY, Zhou CX (2013) MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis 34(3): 713-722.
- Jin H, Li Q, Cao F (2017) miR-124 inhibits lung tumorigenesis induced by K-ras mutation and NNK. molecular therapy. Nucleic acids 9: 145-154.
- Romero Cordoba SL, Salido Guadarrama I, Rodriguez Dorantes M (2014) miRNA biogenesis: biological impact in the development of cancer. Cancer Biol Ther 15(11):1444-1455.
- Xu X, Li S, Lin Y (2013) MicroRNA-124-3p inhibits cell migration and invasion in bladder cancer cells by targeting ROCK1. J Transl Med 11: 276.
- Yuan Q, Sun T, Ye F (2017) MicroRNA-124-3p affects proliferation, migration and apoptosis of bladder cancer cells through targeting AURKA. Cancer Biomark 19(1): 93-101.
- Wang X, Wu Q, Xu B (2015) MiR-124 exerts tumor suppressive functions on the cell proliferation, motility and angiogenesis of bladder cancer by fine-tuning UHRF1. FEBS J 282(22): 4376-4388.
- Zhou W, He L, Dai Y (2018) MicroRNA-124 inhibits cell proliferation, invasion and migration by targeting CAV1 in bladder cancer. Exp Ther Med 16(4): 2811-2820.
- Xiang L, Ou H, Li X (2017) Loss of tumor suppressor miR-126 contributes to the development of hepatitis B virus–related hepatocellular carcinoma metastasis through the upregulation of ADAM9. Tumor Biology 39(6): 1010428317709128.
- Yang J, Lan H, Huang X (2012) MicroRNA-126 inhibits tumor cell growth and its expression level correlates with poor survival in non-small cell lung cancer patients. PLoS One 7(8): e42978.
- Dong Y, Fu C, Guan H (2016) Prognostic significance of miR-126 in various cancers: a meta-analysis. Onco Targets Ther 9: 2547-2555.
- Jia AY, Castillo Martin M, Bonal DM (2014) MicroRNA-126 inhibits invasion in bladder cancer via regulation of ADAM9. Br J Cancer 110(12): 2945-2954.
- Qin A, Wen Z, Zhou Y (2013) MicroRNA-126 regulates the induction and function of CD4+Foxp3+regulatory T cells through PI3K/AKT pathway. J Cell Mol Med 17(2): 252-264.
- Xiao J, Lin HY, Zhu YY (2016) MiR-126 regulates proliferation and invasion in the bladder cancer BLS cell line by targeting the PIK3R2-mediated PI3K/Akt signaling pathway. Oncotarget Ther 9: 5181-5193.
- Zhang HD, Sun DW, Mao L (2015) MiR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem Biophys Res Commun 465(4): 702-713.
- Huang LL, Huang LW, Wang L (2017) Potential role of miR-139-5p in cancer diagnosis, prognosis and therapy. Oncol Lett 14(2): 1215-1222.
- Cao B, Wang K, Liao JM (2016) Inactivation of oncogenic cAMP-specific phosphodiesterase 4D by miR-139-5p in response to p53 activation. Elife 5: e15978.
- Yonemori M, Seki N, Yoshino H (2016) Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci 107(9): 1233-1242.
- Luo H, Yang R, Li C (2017) MicroRNA-139-5p inhibits bladder cancer proliferation and self-renewal by targeting the Bmi1 oncogene. Tumour Biol 39(7): 1010428317718414.

 Research Article
Research Article