Abstract
Several Schiff’s base derivatives for the three structural isomers of phenylenediamine have been designed and evaluated for their DNA intercalation ability. ADMET properties and drug-likeliness of the investigated compounds were predicted by swissADME software. The starting geometry of each investigated compound was constructed using chem3D Ultra software. Molecular docking studies for the intercalation of all compounds with DNA were performed using PyRx software based on Vina as docking software and Lamrkian genetic algorithm as scoring function. The three isomers of phenylenediamine (ortho, meta and para) were used to design the investigated Schiff’s base derivatives, which were divided into several groups (A-G) within two series. Our dockings result of the first series revealed the importance of meta phenylenediamine derivatives over the other (ortho and para) isomers. In addition, they showed the vital role of the naphthyl ring as an aromatic side chain especially when it has a hydroxyl substituent at position 2 of the naphthyl ring. Series 2 contains 4 groups (D-G), and their docking results indicate the importance of another substituent (nitro, methyl, isopropyl, chloro) at position 4 of the aromatic ring. Two main modifications have led to good binding affinities at the DNA intercalatingsite. The first modification involved the use of three ring systems (anthracene or phenanthrene), while the second modification connects the aromatic ring via position 2 instead position 1of the substituted aromatic ring. In conclusion, we have successfully designed several symmetrical meta phenylenediamine Schiff`s bases which can be easily synthesized and used as DNA intercalators. Among all the studied compounds, the derivative which has a hydroxyl anthracene ring connected via position 2 to the meta isomer of phenylenediamine (compound BNG702) showed the highest DNA binding affinity.
Keywords:Schiff`s Base; Phenylenediamine; Dna Intercalation; Molecular Docking; Bioactivity; Antibiotic
Introduction
There are several ways by which organic compounds can interact with Deoxyribonucleic Acid (DNA), including covalent bonding, electrostatic binding or intercalation. Intercalation occurs when the compound (ligand) of appropriate size and chemical nature fit between base pairs of DNAs [1,2]. In order for an intercalator to fit between base pairs, they must consist of planar polycyclic aromatic rings, and the DNA must dynamically open a space between its base pairs by unwinding [3]. The ability of intercalating agents to inhibit DNA replication and nucleic acid synthesis in vivo leads to their extensive use as mutagens, antibiotics, antibacterials, trypanocides, schistosomicides, and antitumor agents. The intercalative binding is non-covalent stacking interaction resulting from the insertion of a planar aromatic ring(s) between the base pairs of the DNA double helix [4,5]. Many classes of organic compounds have been reported to possess good DNA intercalating activity. Among these agents, Schiff’s bases (imines) showed a large range of biological activities including antiviral, antibacterial, antiproliferative, antifungal, and antimalarial activities [6-8].
The vital role of phenylenediamine Schiff bases derivatives as DNA intercalators have been widely described by many research groups. Recently two novel complexes of Schiff bases derived from the condensation of o-phenylenediamine with 3-ethoxysalicylaldehyde have been reported along with their DNA binding, antimicrobial screening and anticancer activities [9,10]. The results demonstrate that the investigated complexes exhibit effective cytotoxicity against growth of carcinoma cells with respect of its imine ligand. Later in 2019, docking studies of three chromium complexes with o-phenylenediamine as secondary ligands with DNA interaction activity have been described [11]. The kinetic parameters revealed that the binding of the complexes with DNA were surface bindings, mainly due to groove binding. Very recently, our research group reported on the DNA interacting activity of some novel symmetrical ortho-phenylenediamine Schiff’s base derivatives. The binding properties of these compounds to genomic DNA (G-DNA) have been investigated indicating that all compounds studied can interact G-DNA [12]. In this work, we aimed to design and predict the DNA intercalating ability of some Schiff’s base derivatives obtained from three structural isomers of phenylenediamine using four different DNA fragments as possible intercalating sites
Materials and Methods
Prediction of Admet Properties and Drug-Likeliness
To verify drug-likeliness, canonical SMILES format of the investigated compounds were used in swissADME (T) online software (http://www.swissadme.ch/index.php) [13]. The main bioavailability parameters were molecular weight (g/mol), lipophilicity (XlogP3), solubility (log S), polarity (Topological Polar Surface Area - TPSA in Å2), saturation (fraction of carbon atoms in Sp3 hybridization – Csp3) and flexibility number of rotatable bonds). Consequently, absorption parameters such as Human Intestinal Absorption, Blood Brain Barrier, P-glycoprotein interaction (substrate or inhibitor) and metabolism (inhibitor or substrate) of the bio-active molecules with different cytochrome P450 enzymes were also estimated using ADMET-SAR, which is based on QSAR data for prediction of Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET)[14,15]. Drug-like molecules considering Lipinski rules and good ADMET properties were chosen as ligands in consequent molecular docking procedures.
Molecular Docking
The starting geometry of all investigated compounds was constructed using Chem3D Ultra (version 12.0, Cambridge soft Com., USA). The optimized geometry of tested compounds with the lowest energy was used in molecular docking studies. Crystal structures of the DNA fragments (1k2j, 1zew, 2des, and 1cp8) were downloaded from the Protein Data Bank (https://www. rcsb.org/structure/102D). Molecular dockings of all compounds with DNA fragments were accomplished by PyRx using Auto Dock 4.2 software from the Scripps Research Institute (TSRI) (http://autodock.scripps.edu/). The dominating configuration of the binding complex of BN compounds and DNA fragments with minimum energy of binding were determined.
Optimization of DNA
The crystal structure of DNA molecules were downloaded from the protein data bank (http://www.rcsb.org/pdb) and their structure analyzed by Discovery Studio Visualizer (http:// accelrys.com). DNA sequences used in the docking studies were: DNA (5’-D(*CP*GP*TP*AP*CP*G)-3’) (PDB ID: 1k2j), DNA (5’-D(*CP*GP*TP*AP*CP*G)-3’) (PDB ID: 1zew), DNA (5’-D(*CP*GP*TP*AP*CP*G)-3’) (PDB ID: 2des), and DNA (5’-D(*CP*GP*TP*AP*CP*G)-3’) (PDB ID: 1cp8). Auto Dock 4.2 software was used to correct imported DNA coordinates (http:// autodock.scripps.edu/). The corrections involved deletion of unwanted water molecules, adding hydrogen atoms (polar only) followed by adding kolloman charge and saved as pdpqt format used as input to PyRx software.
Designing and Preparation of Phenylenediamine Derivatives Compounds
The starting geometry of all investigated compounds was constructed using Chem3D Ultra (version 12.0, Cambridge soft Com., USA) as described in DNA optimization. The optimized geometry of tested compounds with the lowest energy was used in the molecular dockings. In addition, the discovery studio visualizer was used for all tested compounds and converted to PDB files and used as input in PyRx software.
Docking Process and Analysis
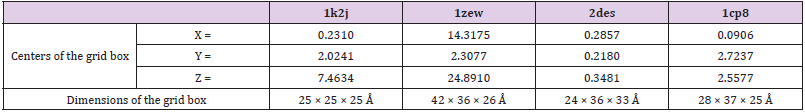
The DNA crystal structure used in this investigation was loaded in PyRx virtual screening tool as pdbqt format. The standard drug and designed compoundspdb files were loaded and automatically converted to pdbqt format. The docking was performed with autodockvina. The centersand the dimensions of the grid box were assigned for each DNA segment as described in Table 1. The best free energy of binding values was obtained and the best interactions were visualized by Discovery Studio.
Table 1: Centers of the grid box and dimensions of the box for DNA fragments (1k2j, 1zew, 2des, and 1cp8).
Results and Discussion
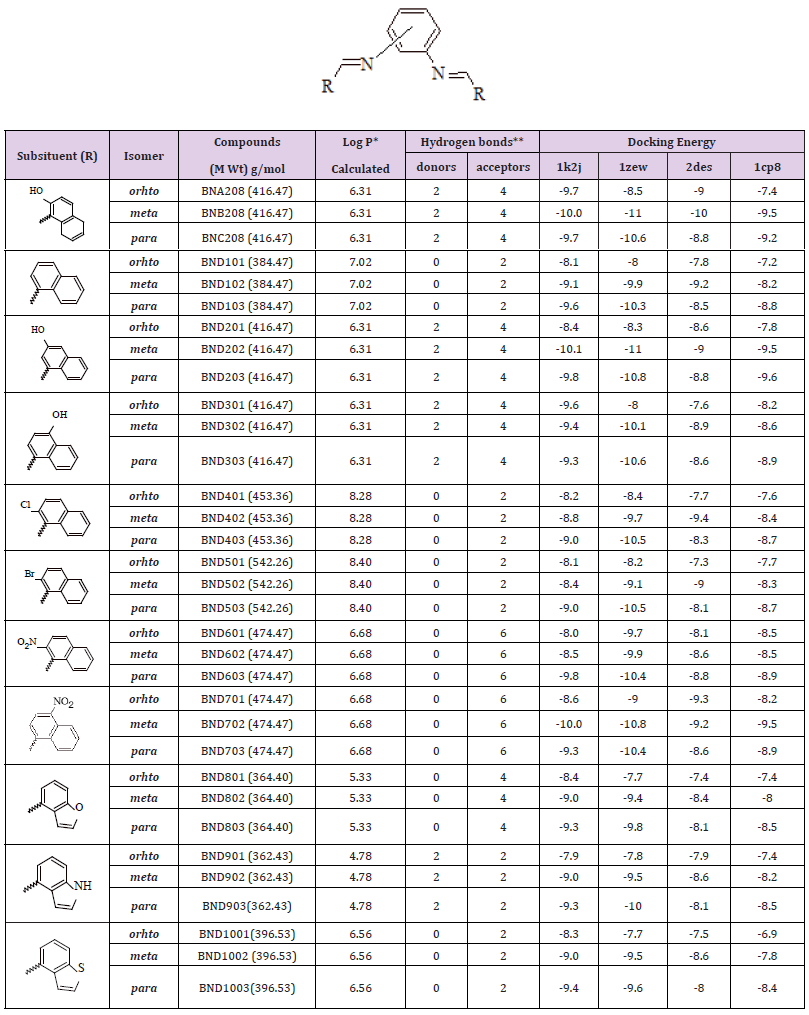
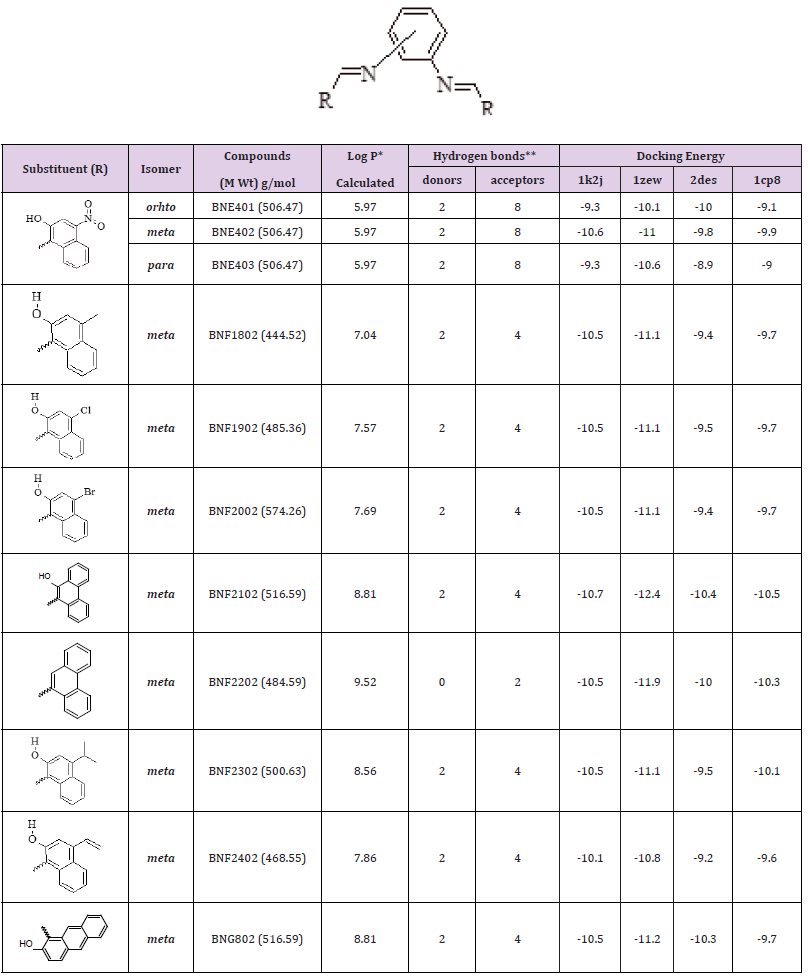
All possible structural isomers of phenylenediamine Schiff’s base derivatives targeted in this study are based on three isomers of phenylenediamine (ortho, meta and para) which were used as main skeleton for the investigated Schiff’s base derivatives. The investigated compounds were divided into two series:
1. in three main groups (A, B and C) for each isomer of phenylenediamine. Each group was subdivided into two subgroups (1 or 2), subgroup 1 indicating the monosubstituted phenylenediamine derivatives, while subgroup 2 indicates the disubstituted phenylenediamine derivatives.
2. several compounds have been designed and tested insilico based on the achieved results from either the first series and/or other compounds of series 2.
The phenylenediamine Schiff’s bases (imines) derivates contain an azomethine group [-HC=N-] in their structure as a result of the condensation of either one or both primary amino groups of phenylenediamine with an aromatic aldehyde. The most important universal structural feature which may play an important role in the DNA intercalating ability is the planar polyaromatic system which inserts between DNA base-pairs with a marked preference for the purine-3’-pyrimidine-5’ sequence [16].
Design Of Phenylenediamine Schiff’s Base Derivatives
Based on the importance of phenylenediamine Schiff’s base derivatives as DNA intercalators as described in the introduction part, and because of their versatile route of chemical synthesis, the three structural isomers of phenylenediamine were used to develop a series of aromatic imino analogues. Comparing the 3-D structures of ortho, meta and para isomers phenylenediamine Schiff’s base derivates indicate that meta and para isomers are more planar than the ortho isomer. This could be attributed to the increased steric interaction of the two imino substituents in ortho positions.
ADMET Properties
The pharmacokinetic properties of all investigated compounds were studied using Lipinski’s rule of five and toxicity. All synthesized compounds were not found to violate the rule. This could be attributed to the rotatable bonds and polar surface areas within the range of maximum permissible limits and thus proving their drug likeness properties. Lipophilicity (LogP o/w) values of all synthesized compounds were in the range from 1.83 to 3.44 for mono-substituted compounds, and 3.10 to 6.31 for disubstituted compounds while their molecular weights are generally within the accepted range- with a few exceptions. The compounds were found with optimum number of hydrogen bonding donors (0-4) and hydrogen bonding acceptors (1-8). The achieved results indicate that most of the compounds follow Lipinski rule and have a good partition coefficient.
Molecular Docking Analysis
In this part of the study, the molecular docking of all compounds with four DNA fragments (1k2j, 1zew, 2des, and 1cp8) were performed by PyRx software using Auto Dock 4.2 to predict the DNA binding ability of the investigated compounds to study their possible binding mode. The molecular docking results showed that van der Waals and hydrogen bonding are mainly involved in the interaction between the tested compounds and the DNA fragments. The contribution of van der Waals and hydrogen bonding interaction is much greater than that of the electrostatic interaction because the sum of van der Waals energy, hydrogen bonding energy and desolvation free energy is larger than the electrostatic energy. The DNA fragment (1k2j) was used to discuss and compare the intercalating activity of all compounds. It appears worth to note that compounds which exhibit high activity against (1k2j) also showed high activity against the other DNA fragments. Simultaneously, any modification which might decrease the activity against (ik2j) will produce the same effect against all other DNA fragments.
Reference Compounds
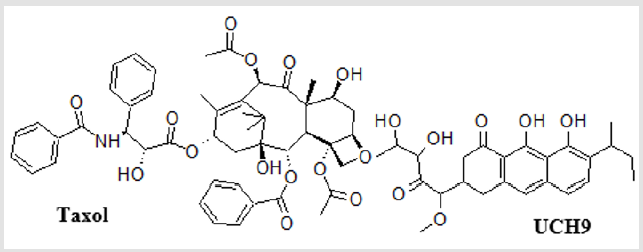
Two well-known biologically active DNA intercalating agents were used as references (Figure 1), namely paclitaxel (taxol) and the aureolic acid group antitumor drug, UCH9. Taxol is considered a successful drug in the treatment of various forms of cancers [17- 19]. UCH9 contains more hydrophilic parts (polyhydroxyl groups) and longer hydrophobic side chains (aromatic rings). Both of the compounds bind to the minor groove of DNA and cause DNA unwinding. The docking energy of both reference compounds against the tested DNA fragments is depicted in Table 2.
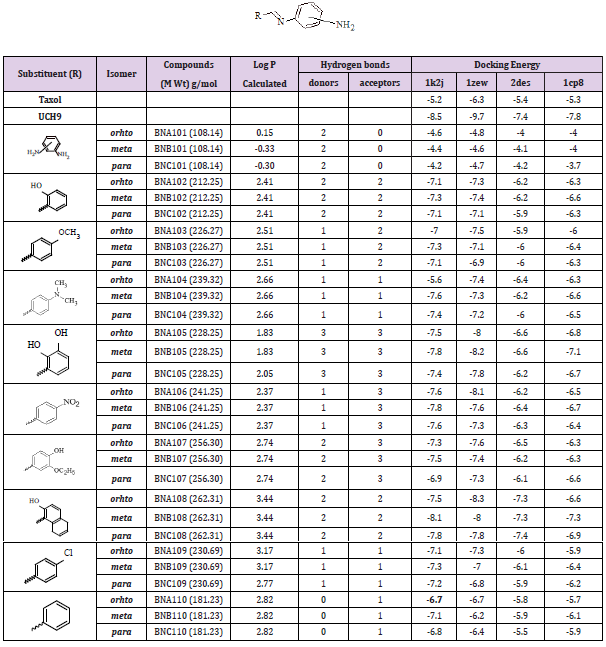
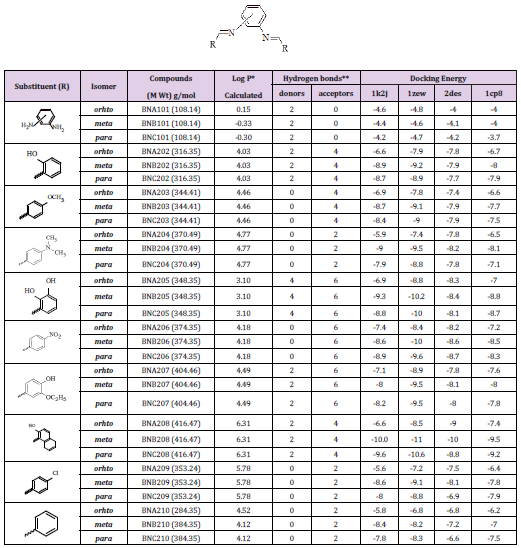
Series (I): Monosubstituted (groups A1, B1, and C1) and disubstituted (groups A2, B2, and C2) phenylenediamine derivatives were designed based on the substitution of three structural isomers (ortho, meta, and para) phenylenediamine with several aromatic aldehydes, where the aldehydes are differ in their lipophilicity, hydrogen bond ability as donor or acceptors, and ionizability. The chemical structures, partition coefficient, hydrogen bond ability, and docking energy of the monosubstituted and disubstituted derivatives are listed in Tables 2 & 3, respectively.
Table 2: Various energies in the binding process of the monosubstituted phenylenediamine derivatives (group A1, B1, and C1) with DNA obtained from molecular docking. Units of energies are in kcal/mol.
Table 3: Various energies in the binding process of the disubstituted phenylenediamine derivatives (group A2, B2, and C2) with DNA obtained from molecular docking. Units of energies are kcal/mol.
Discussion of Docking Results of Series I Compounds
Based on the results of series I phenylenediamine Schiff’s base derivatives, we predict an increased DNA intercalating efficacy for all series I derivatives. This prediction was confirmed since the docking energy of all derivatives are less than the corresponding phenylenediamine isomer, which may be attributed to the introduction of extra aromatic rings to the unsubstituted phenylenediamine isomer. In general, the di-substituted isomer of phenylenediamine is more active than the corresponding monosubstituted isomer except for ortho isomers, which is probably due to steric obstruction affecting the planarity of the three aromatic rings (Figure 2). In addition, meta isomers are the most active among the three isomers, whereas para isomers are generally more active than ortho isomers. Introduction of hydrophilic substitutes (R) to the aromatic ring of either monosubstituted or di-substituted derivatives leads to a decrease of the intercalating efficacy, viz. an increase of the docking energy. Whereas the introduction of hydrophobic substitutes (R) to the same aromatic ring leads to an increase of the intercalating efficacy, viz. a decrease of the docking energy. The highest efficacy was predicted for an introduction of 2-hydroxynaphthaldehyde, such as BNA108, BNA208, BNB108, BNB208, BNC108, and C208 compounds. Increasing the hydrogen bond donor ability of the aromatic ring substitutes (R) of either monosubstituted or disubstituted derivatives leads to increase of the intercalating efficacy, viz, a decrease of the docking energy, as can see from the comparison of monohydroxy phenyl derivatives (102 / 202 for all groups) with nonsubstituted derivatives (110 / 210 for all groups).
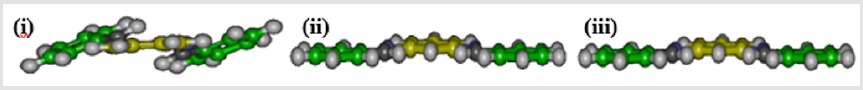
Figure 2: Planarity of the three aromatic rings of meta (ii) and para (iii) isomers in comparison to ortho (i) isomers of compound 210.
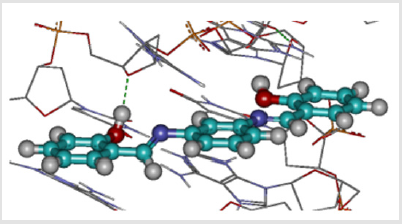
From results depicted in Tables 2 & 3, we infer the importance of hydroxyl substituents at the ortho positions of the aromatic ring, as it leads to an increase of intercalating efficacy. The highest efficacy was found with meta phenylenediamine isomers, whereas ortho isomers show the lowest efficacy which could be related to the planarity of the molecules as explained above. For di-substituted derivatives, only one hydroxyl group may form hydrogen bonds with the DNA as explained in Figure 3 which shows hydrogen bonding of compound BNA202 with DNA. Introduction of a second hydroxyl group at meta position to the hydroxyphenyl ring has no effect on the DNA intercalating efficacy except for compound BNB202, in which such a modification may lead to a slight improvement of its DNA intercalating efficacyas shown with BNB205.
Figure 3: Compound BNB202 interacting between the two DNA strands with hydroxyl groups (red) as hydrogen bond donors.
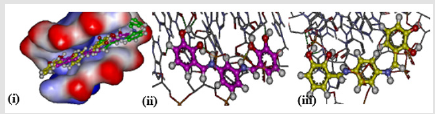
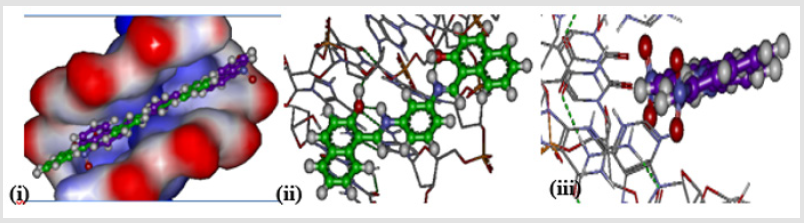
Comparing the localization of the three isomers for all derivatives at the intercalating site indicated that both meta and para derivatives could be localized nearly at the same binding site, while ortho isomers bind at a different site. This could be attributed to the non-planarity of the ortho isomer (Figure 4(i)). The possible difference in the binding ability of meta and para isomers could be attributed to the number of hydrogen bonds which are formed at intercalating sites, where the small increase in the efficacy of the meta isomer (BNB205) appears related to formation of two hydrogen bonds, in addition to keeping the planarity which is needed for optimum localization at intercalating sites as shown in Figure 4(ii). The third isomer (para isomer) could form only one hydrogen bond at the intercalating site since only one hydroxyl group can interact with the DNA as shown in Figure 4(iii). Introduction of a hydroxyl group at the para position of the aromatic ring leads to a decrease of the intercalation efficacy if compared to other hydroxyl derivatives (BNB202 and BNC202). Even though, ortho isomers show a slight increase in efficacy (BNA207) because it improves the planarity and consequently the localization at efficient sites in comparison to the compound (BNA202).
Figure 4: (i) Compounds BNA205 (green), BNB205 (purple), and BNC205 (yellow) at the intercalating site. (ii) Hydrogen bonds formed by BNB205. (iii) Hydrogen bonds formed by BNC205.
Introduction of a nitro group at para position at the phenyl group leads to slight increase in the intercalating efficacy. This could be easily inferred from the comparison of BNA206, BNB206, and BNC206 with the corresponding non-substituted derivatives. The slight increase in efficacy may be related to the hydrogen bond acceptor ability of the oxygen atom in the nitro group in all p-nitro derivatives. At the same time, the localization of all nitro derivatives is different compared to the other compounds. The highest DNA intercalating efficacy of series I is compound BNB208, which has an extra phenyl moiety attached to the first phenyl substituent of the hydroxyl analogue of the meta isomer phenylenediamine Schiff`s base derivative. The produced naphthyl group with the hydroxyl substituent has the advantage of both hydrogen bonds forming derivatives (as it has hydroxyl groups) and the high lipophilic character as result of its increased lipophilicity (high log P value because of the naphthyl ring).
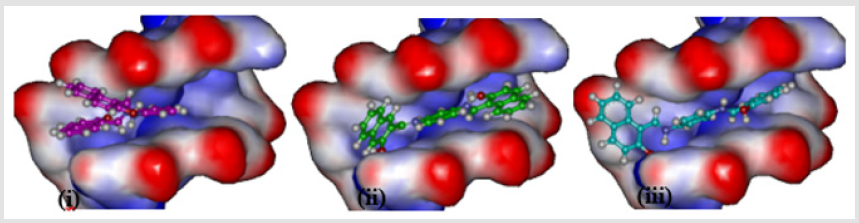
Comparing the localization of the three isomers of di-2- hydroxynaphthyl analogues (ortho (BNA208), meta (BNB208), and para (BNC208)), as shown in Figure 5, indicate that both meta and para isomers which show increased efficacy, when compared with ortho isomers, are located at the same location within the intercalating site (minor groove of DNA), whereas the ortho isomer has a twisted shape as result of its non-planarity. Investigation of the interaction of BNB2028 (meta isomer) with the DNA intercalating site shows that only one of the two hydroxyl groups is involved in formation of a hydrogen bond with the DNA nucleotide as donor. The other hydroxyl group is involved in intramolecular hydrogen bonding with the azomethine group.
Figure 5: Localization of (i) BNA208, (ii) BNB208, and (iii) para BNC208 at the intercalating site of DNA.
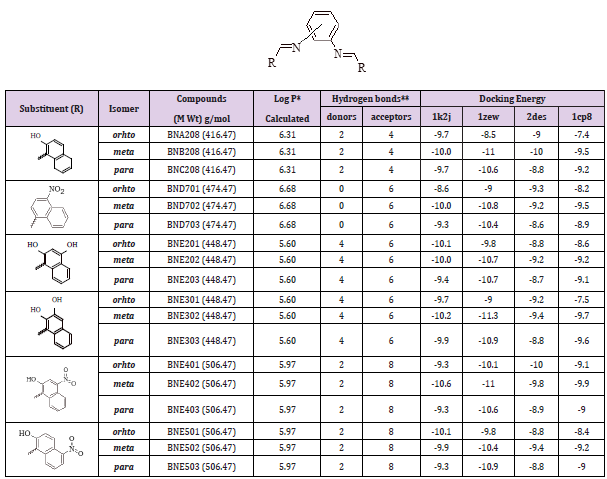
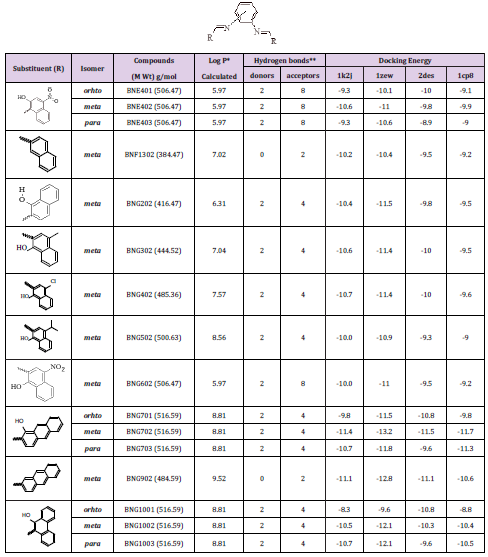
Series (II): Only disubstituted isomers ofphenylenediamine were investigated in our study, because the results obtained from series I compounds indicated that they are more active than the corresponding monosubstituted isomers. In this series, the most active derivative from each group was used as a lead structure to design the following group to develop more active analogues.
Group D: Disubstituted Naphthyl Phenylenediamine Derivatives
Group D derivatives were designed based on the docking results of series 1 group, which indicated the important role of the naphthyl group where an aromatic side chain, several hydrophilic (OH, NO2) and hydrophobic (Cl, Br) substituents were added. On the other hand, some of these groups are hydrogen bond donors while the others are only acceptors. The third part of these substituents are non-hydrogen bonding groups. The possible role of heterocyclic analogues of the naphthyl group were investigated using indole, benzofuran and benzothiophene rings. Chemical structures, partition coefficient, hydrogen bond ability and docking energies of group D derivatives are listed in Table 4.
Table 4: Various energies in the binding process of the disubstituted naphthyl phenylenediamine derivatives (group D) with DNAs obtained from molecular docking. The unit of all energies are kcal/mol.
Discussion of Docking Results of Group D Compounds
Comparing the docking energies of group D compounds indicates that the hydroxyl substituent on the naphthyl side chain is important for activity, it could be at positions 2, 3, or 4 of the naphthyl ring with optimum activity at positions 2 or 3 of meta isomers. Hydrogen bond acceptor substituent (NO2) at position 2 of the naphthyl side chain leads to a small decrease in the intercalating activity when compared with the non-substituted derivatives. Whereas the same substituent at position 4 of the naphthyl group leads to large increase in the activity of meta isomers. Lipophilic substituents (Cl or Br) at position 2 of the naphthyl side chain had no effect on the intercalating activity when compared with the non-substituted derivative (BND 101 – 103). The same effect was obtained upon the replacement of the naphthyl group by heterocyclic rings (indole, benzofuran or benzothiophene rings). Investigation of binding mode of BNB208 and BNB702 showed that both compounds have a different localization as described in Figure 6(i). The hydroxyl group in BNB208 interacts with only one hydrogen bond (Figure 6(ii)), while the nitro group in BNB702 forms no hydrogen bond with a DNA binding site (Figure 6(iii)), but it may lead to an optimum localization in an active site. In conclusion, the docking results achieved for this group indicates that hydroxyl and nitro substituents on the naphthyl side chain at positions (2 (BNB208) or 4 (BND202)) and (4 (BND702), respectively, will result in optimum DNA intercalating activity.
Figure 6: (i) Localization of BNB208 (green) and BND702 (blue) at the intercalating site of DNA. (ii)Compound BNB208 interacting with DNA strands with one hydroxyl group as intermolecular hydrogen bond donor. (iii) Localization of dinitro derivative BND702 (purple) in comparison to hydroxyl derivative (BNB202, brown).
Group E: Disubstituted Hydroxyl and Nitro Naphthyl Phenylenediamine Derivatives
Group E compounds were designed based on disubstitution of the three structural isomers (ortho, meta, and para) phenylenediamine with several hydroxy and/or nitro naphthaldehydes, where the aldehydes differ in their position on the naphthyl ring. The chemical structures, partition coefficient, hydrogen bond ability and docking energies of group D derivatives are listed in Table 5.
Table 5: Various energies in the binding process of disubstituted hydroxy and/or nitro naphthyl phenylenediamine derivatives with DNA obtained from molecular docking. The unit of all energies are kcal/mol.
Discussion of Docking Results of Group E Compounds
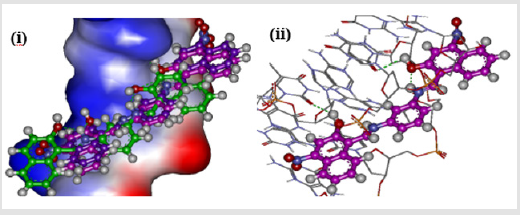
The possible positional isomers of either hydroxyl and/or nitro naphthyl side chain were targeted in the design of group E compounds. Comparing the docking energies of all compounds indicated that positions 2 and 4 of the naphthyl group are the best locations of both hydroxyl and nitro substituent as in compound BNE402, which are localized at the same position of the DNA intercalating site as the nitro derivative BND702 (different to compound BNB208) as described in Figure 7(i). The two hydroxyl groups of compound BNE402 could interact with the hydrogen bond as donor (instead of only one hydroxyl as for compound BNB208), while both nitro groups may locate the compound in suitable conformation at the active site and have no hydrogen bonding with the DNA (Figure 7(ii)).
Figure 7: (i) Localization of BNB208 (green), BND702 (blue), and BNE402 (purple) at the intercalating site of DNA. (ii)Compound BNE402 interacting with DNA strands with both hydroxyl groups as intermolecular hydrogen bond donor.
Group F: Lipophilic Analogues of Disubstituted Hydroxyl and Nitro Naphthyl Phenylenediamine Derivatives
The strategy which applied to design this group was based on substitution of nitro groups of compound BNE402 (the most active of group E) with non-hydrogen bonding lipophilic groups. In addition, the effect of replacing of the naphthyl ring with a three ring system (anthracene or phenantrene) was also investigated. The chemical structures, partition coefficient, hydrogen bond ability and docking energies of group F derivatives are listed in Table 6.
Table 6: Various energies of the binding process of the lipophilic disubstituted hydroxy and/or nitro naphthyl meta isomer of phenylenediamine derivatives with DNA obtained from molecular docking. The unit of all energies are kcal/mol.
Discussion of Docking Results of Group E Compounds
Most of the compounds tested in this group showed increased ability to intercalate with the DNA as their docking energies are close to 10.5. This indicated that we use a suitable strategy to develop improved analogues based on previous groups. The achieved docking results indicated that all substituents used in this group have the same effect on the location of the produced compounds at the intercalating site and have nearly the same potency in comparison to compound BNE402. The only exception is compound BNF2402, which contains a vinyl substituent on the para position at the naphthyl side chain. This finding was concluded by comparison of compound BNE402 with the isopropyl analogue BNF2302, where both of them occupy the same part of the intercalating site at the DNA in contrast to the vinyl analogue BNF2402 which has a different localization with a decreased binding ability with the active site, as shown in Figure 7(i).
Highly increased activity was predicted with derivatives which have anthracene or phenantrene rings replacing the naphthyl ring. Substitution of the naphthyl group with a phenantrene ring (BNF2102) keeps the compound within the same partused by BND802, as explained in Figure 8(ii). Addition of a hydroxyl group at the ortho position of the phenantrene ring may slightly increase the binding interaction. Substitution of the anthracene ring in place of the naphthyl ring (BNG802) may locate the compound in a different location in comparison to the phenantrene derivatives with nearly the same activity as shown in Figure 8(ii).
Figure 8: (i) Localization of BNE402 (green), BNF2302 (sky blue), and BNF2402 (purple) at the intercalating site of DNA. (ii)Localization of BNE402 (green), BNF2102 (orange), BNF2202 (yellow), and BNG802 (purple) at the intercalating site of DNA.
Group G: Lipophilic Analogues of Disubstituted Hydroxyl and Nitro Naphth-2-Yl-Phenylenediamine Derivatives
Group G was designed based on the disubstitution of the meta isomer of phenylenediamine with several substituted 2-naphthaldehydes in place of the previously used 1- naphthaldehydes.This modification was postulated to investigate the effect of varying the connection of the naphthyl ring from position 1 to position 2. The substituents which showed optimum intercalation ability with DNA in previous groups were used in different positions on the naphthyl group. In addition, substitution of the naphthyl ring with three ring system (anthracene or phenantrene rings) were also investigated. The chemical structures, partition coefficient, hydrogen bond ability, and docking energies of group F derivatives are listed in Table 7.
Discussion of Docking Results of Group G Compounds
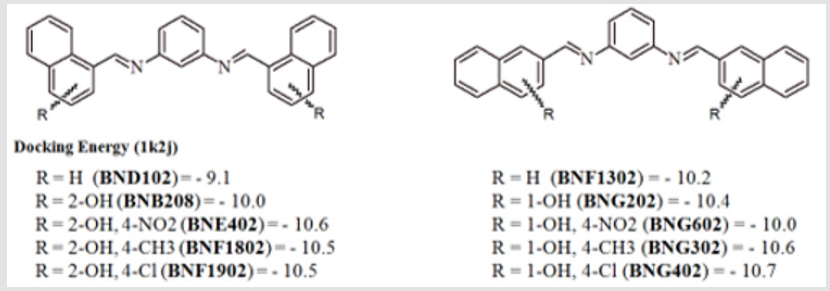
This group of compounds showed highly increased ability to intercalate with the DNA in comparison to previous groups investigated. Substitution of 1-naphthyl (BND102) with 2-naphthyl (BNF1302) slightly increased the DNA binding ability. The same effect was achieved after addition of a hydroxyl group as in compounds 1-naphthyl (BNB208) resulting in 2-naphthyl (BNG202). The docking energy was decreased if compound BNE402, which contains a 2-hydroxyl group and a 4-nitro group, has been modified to generate the corresponding 2-naphthyl analogue, The analogues which contain 4-methyl or 4-chloro (lipophilic) groups have nearly equal activities for 1-naphthyl and2-naphthyl derivatives as summarized in Figure 9.The main difference in the DNA intercalation of both 1-naphthyl and 2-naphthyl derivatives can be \attributed to the difference in the localization within the intercalating site, where 2-naphthyl derivatives occupy a larger space than 1-naphthyl derivatives.
Figure 9: Docking energy of some 1-naphthyl and 2-naphthyl phenylenediamine Schiff`s base derivatives.
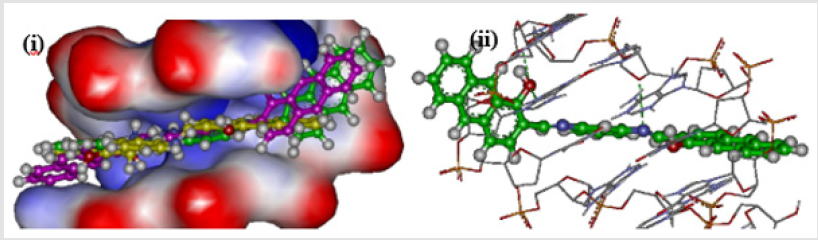
The most effective compound in our study was BNG702, in which meta phenylenediamine is used to produce the corresponding Schiff`s base derivative via condensation with 1-hydroxy anthracene- 2-carboxaldehyde.bThe 3D structure of compound BNG702 showed that the benzene ring of phenylenediamine and one of the anthracene rings are more planar, while the second anthracene ring is perpendicular position (Figure 10). The ortho and para isomers of BNG702 exhibit higher docking energies and consequently they were less efficient as DNA intercalators. The decrease in efficacy of both ortho (BNG701) and para (BNG703) isomers appear to be related to the position occupied by each compound at the active site as described in Figure 10 (i).Investigation of BNG702- DNA intercalation indicated that hydroxyl groups attached to the perpendicular anthracene ring could form hydrogen bonds with DNA nucleotides. The other hydroxyl group appears not to be involved in such binding with the target as shown in Figure 10 (ii). The nitrogen atom attached to the planer anthracene ring may get involved in hydrogen bond as acceptor.
Figure 10: (i) Localization of ortho (BNG701 (yellow)), meta (BNG702 (green)), and para (BNG703 (sky blue)) isomers of 1-hydroxy anthracene-2- derivatives of phenylenediamine at the intercalating site of DNA. (ii) Intermolecular hydrogen bonds of the most active derivative (BNG702) at the intercalating site of DNA.
Our docking results are consistent with five Schiff base ligands that were synthesized and fully spectroscopically characterized. The five Schiff base ligands which contain stronger electron donating substituents had higher DNA-binding ability than the nonelectron donating analogues [20]. The molecular docking results of the five Schiff base ligands showed that all of the synthesized ligands were minor groove binders and the H-bond interactions play a dominant role in the stability of ligand-DNA [20]. In addition, another binding study of a series of p-dimethylaminobenzaldehyde Schiff bases derivatives with Ct-DNA was performed using absorption spectroscopy, viscosity measurement, circular dichroic, molecular docking employing B-DNA dodecamer d(CGCGAATTCGCG)2, fluorescence quenching and molecular dynamics simulation methods [21]. The synthesized Schiff bases were tested for antimicrobial activity against bacterial species Klebsiella pneumoniae Escherichia coli, Staphylococcus aureus and Pseudomonas aeruginosa, using the disc diffusion method. All the tested Schiff bases compounds showed promising effects against the tested bacterial strains [21]. Two previous studies are consistent with our results which renders our designed compounds promising drugs against some tumours and bacterial strains.
Conclusions
We have successfully performed virtual screening for all structural isomers of symmetrical phenylenediamine Schiff`s basederivatives with four DNA fragments. They are good candidates for the development of DNA intercalators. The highest efficacy among the three isomers of phenylenediamines where shown by meta isomers. Hydroxyl groups at the ortho position of the aromatic phenyl ring of all series play an important role in compound- DNA interaction. Moreover, addition of a second phenyl ring which produces high efficacy of the naphthyl side chain as in compound BNB208 is considered a key compound developed from series 1 compounds. Variation of both substituents and the position of the linking of the naphthyl group to the meta isomer of phenylenediamine via imine functionality from position 1 to position 2, leads to an even higher increase in efficacy of many derivatives. Interestingly, the same positional variation of the connection between the hydroxy anthracene ring and the meta isomer of phenylenediamine shows the most potent DNA intercalating efficacy achieved in the current study (compound BNG702), which is, therefore, considered the most promising lead compound in the production of potent DNA-intercalators. In conclusion, we highly recommend the synthesis and in vitro testing of meta phenylenediamine Schiff`s base derivatives with two rings (naphthyl) together with a three (anthracene) ring system.
References
- Plavec J, Kocman V, Kotar A (2019) Intercalation of heterocyclic ligand between quartets in G-rich tetrahelical structure. Chemistry.
- Barrett S, De Franco M, Kellett A, Dempsey E, Marzano C, et al. (2019) Anticancer activity, DNA binding and cell mechanistic studies of estrogen-functionalised Cu(II) complexes. J Biol Inorg Chem.
- Khan TA, Naseem S, Khan SN, Khan AU, Shakir M (2009) Synthesis and spectral characterization of 14- and 16-membered tetraazamacrocyclic Schiff base ligands and their transition metal complexes and a comparative study of interaction of calf thymus DNA with copper (II) complexes. Spectrochim Acta A Mol BiomolSpectrosc 73(4):622-629.
- Bosire R, Nanasi PJr, Imre L, Dienes B, Szoor A, et al. (2019) Intercalation of small molecules into DNA in chromatin is primarily controlled by superhelical constraint. PLoS One 14(11):e0224936.
- Bunkenborg J, Gadjev NI, Deligeorgiev T, Jacobsen JP (2000) Concerted intercalation and minor groove recognition of DNA by a homodimericthiazole orange dye. Bioconjug Chem 11(6):861-867.
- Rakesh KP, Kumara HK, Ullas BJ, Shivakumara J, Channe GD (2019) Amino acids conjugated quinazolinone-Schiff's bases as potential antimicrobial agents: Synthesis, SAR and molecular docking studies. Bioorg Chem 90:103093.
- Wu Y, Li Q, Zhang X, Li Y, Li B, et al. (2019) Cellulose-based peptidopolysaccharides as cationic antimicrobial package films. Int J Biol Macromol 128:673-680.
- Sabaa MW, Elzanaty AM, Abdel-Gawad OF, Arafa EG (2018) Synthesis, characterization and antimicrobial activity of Schiff bases modified chitosan-graft-poly(acrylonitrile). Int J Biol Macromol 109:1280-1291.
- Shahabadi N, Momeni BZ, Zendehcheshm S (2019) Studies on the Interaction of [SnMe2Cl2(bu2bpy)] Complex with ct-DNA Using Multispectroscopic, Atomic Force Microscopy (AFM) and Molecular Docking. Nucleosides Nucleotides Nucleic Acids 38(2):157-182.
- Chakraborty A, Panda AK, Ghosh R, Biswas A (2019) DNA minor groove binding of a well known anti-mycobacterial drug dapsone: A spectroscopic, viscometric and molecular docking study. Arch Biochem Biophys665:107-113.
- P V, SK B, B S, AL PV (2018) Copper-metformin ternary complexes: Thermal, photochemosensitivity and molecular docking studies. Mater Sci Eng C Mater Biol Appl 90:621-633.
- Mohamed SS,Sadawe IA, Meiqal NH, Alshoushan AA, Aboud AS, et al. (2019) DNA Interaction Study of Some Symmetrical 1,2-Phenylenediamine Schiff's Base Derivatives as New Potential DNA Intercalators Using Ethidium Bromide Competition Fluorescent Assay. Arch Biomed Eng & Biotechnol 2(2):1-7.
- Daina A, Michielin O, Zoete V (2017)SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717.
- Guan L, Yang H, Cai Y, Sun L, Di P, et al. (2019) ADMET-score - a comprehensive scoring function for evaluation of chemical drug-likeness. Medchemcomm 10(1):148-157.
- Yang H, Lou C, Sun L, Li J, Cai Y, et al. (2019) admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 35(6):1067-1069.
- Thies H, Schonenberger H, Bauer KH (1958) Reactions of Schiff's bases. V. Transformation of benzylidene-arylamines with mixtures of magnesium and magnesium iodide. Arch Pharm Ber Dtsch Pharm Ges 291/63(11-12):620-627.
- Geromichalos GD, Tarushi A, Lafazanis K, Pantazaki AA, Kessissoglou DP, et al. (2018)In vitro and in silico study of the biological activity of manganese(III) inverse-[9-MC-3]-metallacrowns and manganese(II) complexes with the anti-inflammatory drugs diclofenac or indomethacin. J InorgBiochem187:41-55.
- Karthikeyan S, Hoti SL, Nazeer Y, Hegde HV (2016)Glaucarubinone sensitizes KB cells to paclitaxel by inhibiting ABC transporters via ROS-dependent and p53-mediated activation of apoptotic signaling pathways. Oncotarget 7(27):42353-42373.
- Liu Y, Yang Z, Du J, Yao X, Zheng X, et al. (2008) Interaction of Taxol with intravenous immunoglobulin: An inhibition of Taxol from crystallizing in aqueous solution. Int Immunopharmacol 8(3):390-400.
- Jamshidvand A, Sahihi M, Mirkhani V, Moghadam M, Mohammadpoor-Baltork I, et al. (2018) Studies on DNA binding properties of new Schiff base ligands using spectroscopic, electrochemical and computational methods: Influence of substitutions on DNA-binding. Journal of Molecular Liquids 253:61-71.

 Research Article
Research Article