Short Communication
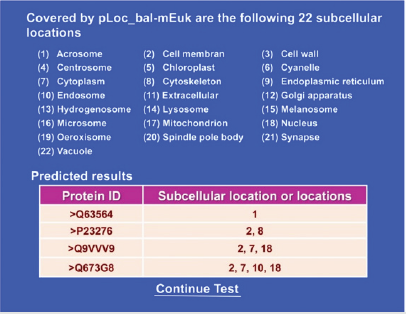
Recently, a very powerful web-server predictor has been established for identifying the subcellular localization of a protein based on its sequence information alone for the multi-label systems [1], in which a same protein may occur or move between two or more location sites and hence needs to be marked with the multilabel approach [2-103]. The web-server predictor is called “pLoc_ bal-mEuk”, where “bal” means the web-server has been further improved by the “balance treatment” [3-9], and “m” means the capacity able to deal with the multi-label systems. To find how the webserver is working, please do the following. Click the link at http://www.jci-bioinfo.cn/pLoc_bal-mEuk/, the top page of the pLoc_bal-mEuk webserver will appear on your computer screen, as shown in (Figure 1). Then by following the Step 2 and Step 3 in [5], you will see the predicted results shown on (Figure 2). Nearly all the success rates achieved by the web-server predictor for the eukaryotic proteins in each of the 22 subcellular locations are within the range of 90-100%, which is far beyond the reach of any of its counterparts. Besides, the web-server predictor has been developed by strictly observing the guidelines of “Chou’s 5-steps rule” and hence have the following notable merits (see, e.g., [10-90] and three comprehensive review papers [2,91,92]):
Figure 2: A semi screenshot for the webpage obtained by following Step 3 of Section 3.5 (Adapted from [5]).
1. Crystal clear in logic development,
2. Completely transparent in operation,
3. Easily to repeat the reported results by other investigators,
4. With high potential in stimulating other sequenceanalyzing methods, and
5. Very convenient to be used by most experimental scientists.
For the fantastic and awesome roles of the “5-steps rule” in driving proteome, genome analyses and drug development, see a series of recent papers [2, 92-104] where the rule and its wide applications have been very impressively presented from various aspects or at different angles
References
- Chou KC, Shen HB (2007) Recent progresses in protein subcellular location prediction. Analytical Biochemistry 370: 1-16.
- Chou KC (2019) Advance in predicting subcellular localization of multi-label proteins and its implication for developing multi-target drugs. Current Medicinal Chemistry 26: 4918-4943.
- Xiao X, Cheng X, Chen G, Mao Q, Chou KC (2019) pLoc_bal-mVirus: Predict Subcellular Localization of Multi-Label Virus Proteins by Chou's General PseAAC and IHTS Treatment to Balance Training Dataset. Med Chem 15(5): 496-509.
- Xiao X, Cheng X, Chen G, Mao Q, Chou KC (2019) pLoc_bal-mGpos: predict subcellular localization of Gram-positive bacterial proteins by quasi-balancing training dataset and PseAAC. Genomics 111: 886-892.
- Chou KC, Cheng X, Xiao X (2019) pLoc_bal-mEuk: predict subcellular localization of eukaryotic proteins by general PseAAC and quasi-balancing training dataset. Med Chem 15(5): 472-485.
- Chou KC, Cheng X, Xiao X (2019) pLoc_bal-mHum: predict subcellular localization of human proteins by PseAAC and quasi-balancing training dataset Genomics 111(6): 1274-1282.
- Cheng X, Lin WZ, Xiao X,Chou KC (2019) pLoc_bal-mAnimal: predict subcellular localization of animal proteins by balancing training dataset and PseAAC. Bioinformatics 35(3): 398-406.
- Cheng X, Xiao X, Chou KC (2018) pLoc_bal-mPlant: predict subcellular localization of plant proteins by general PseAAC and balancing training dataset Curr Pharm Des 24: 4013-4022.
- Cheng X, Xiao X, Chou KC (2018) pLoc_bal-mGneg: predict subcellular localization of Gram-negative bacterial proteins by quasi-balancing training dataset and general PseAAC. Journal of Theoretical Biology 458: 92-102.
- Butt AH, Khan YD (2018) Prediction of S-Sulfenylation Sites Using Statistical Moments Based Features via Chou's 5-Step Rule. International Journal of Peptide Research and Therapeutics (IJPRT).
- Awais M, Hussain W, Khan YD, Rasool N, Khan SA, et al. (2018) iPhosH-PseAAC: Identify phosphohistidine sites in proteins by blending statistical moments and position relative features according to the Chou's 5-step rule and general pseudo amino acid composition. IEEE/ACM Trans Comput Biol Bioinform.
- Barukab O, Khan YD, Khan SA, Chou KC (2019) iSulfoTyr-PseAAC: Identify tyrosine sulfation sites by incorporating statistical moments via Chou's 5-steps rule and pseudo components Current Genomics.
- Butt AH, Khan YD (2019) Prediction of S-Sulfenylation Sites Using Statistical Moments Based Features via Chou's 5-Step Rule. International Journal of Peptide Research and Therapeutics (IJPRT).
- Chen Y, Fan X (2020) Use of Chou's 5-Steps Rule to Reveal Active Compound and Mechanism of Shuangshen Pingfei San on Idiopathic Pulmonary Fibrosis. Current Molecular Medicine.
- Du X, Diao Y, Liu H, Li S (2019) MsDBP: Exploring DNA-binding Proteins by Integrating Multi-scale Sequence Information via Chou's 5-steps Rule. Journal of Proteome Research 18: 3119-3132.
- Ehsan A, Mahmood MK, Khan YD, Barukab OM, Khan SA (2019) iHyd-PseAAC (EPSV): Identify hydroxylation sites in proteins by extracting enhanced position and sequence variant feature via Chou's 5-step rule and general pseudo amino acid composition. Current Genomics 20(2): 124-133.
- Hussain W, Khan SD, Rasool N, Khan SA, Chou KC (2019) SPalmitoylC-PseAAC: A sequence-based model developed via Chou's 5-steps rule and general PseAAC for identifying S-palmitoylation sites in proteins. Anal Biochem 568: 14-23.
- Hussain W, Khan YD, Rasool N, Khan SA, Chou KC (2019) SPrenylC-PseAAC: A sequence-based model developed via Chou's 5-steps rule and general PseAAC for identifying S-prenylation sites in proteins. J Theor Biol 468: 1-11.
- Ju Z, Wang SY (2019) Prediction of lysine formylation sites using the composition of k-spaced amino acid pairs via Chou's 5-steps rule and general pseudo components. Genomics
- Kabir M, Ahmad S, Iqbal M, Hayat M (2019) iNR-2L: A two-level sequence-based predictor developed via Chou's 5-steps rule and general PseAAC for identifying nuclear receptors and their families. Genomics.
- Khan ZU, Ali F, Khan IA, Hussain Y, D Pi (2019) iRSpot-SPI: Deep learning-based recombination spots prediction byincorporating secondary sequence information coupled withphysio-chemical properties via Chou's 5-step rule and pseudo components. Chemometrics and Intelligent Laboratory Systems (CHEMOLAB) 189: 169-180.
- Lan J, Liu J, Liao C, Merkler DJ, Han Q, et al. (2019) A Study for Therapeutic Treatment against Parkinson’s Disease via Chou's 5-steps Rule. Current Topics in Medicinal Chemistry.
- Le NQK (2019) iN6-methylat (5-step): identifying DNA N (6)-methyladenine sites in rice genome using continuous bag of nucleobases via Chou's 5-step rule. Mol Genet Genomics 294: 1173-1182.
- Le NQK, Yapp EKY, Ho QT, Naga Sundaram N, Ou YY, et al. (2019) iEnhancer-5Step: Identifying enhancers using hidden information of DNA sequences via Chou's 5-step rule and word embedding. Anal Biochem 571: 53-61.
- NQK Le, EKY Yapp, YY Ou, HY Yeh (2019) iMotor-CNN: Identifying molecular functions of cytoskeleton motor proteins using 2D convolutional neural network via Chou's 5-step rule. Anal Biochem 575: 17-26.
- Liang R, Xie J, Zhang C, Zhang M, Huang H, et al. (2019) Identifying Cancer Targets Based on Machine Learning Methods via Chou's 5-steps Rule and General Pseudo Components. Current Topics in Medicnal Chemistry.
- Liang Y, Zhang S (2019) Identifying DNase I hypersensitive sites using multi-features fusion and F-score features selection via Chou's 5-steps rule. Biophys Chem 253: 106227.
- Malebary SJ, Rehman MSU, Khan YD (2019) iCrotoK-PseAAC: Identify lysine crotonylation sites by blending position relative statistical features according to the Chou's 5-step rule. PLoS One 14: e0223993.
- Nazari I, Tahir M, Tayari H, Chong KT (2019) iN6-Methyl (5-step): Identifying RNA N6-methyladenosine sites using deep learning mode via Chou's 5-step rules and Chou's general PseKNC. Chemometrics and Intelligent Laboratory Systems (CHEMOLAB).
- Ning Q, Ma Z, Zhao X (2019) dForml(KNN)-PseAAC: Detecting formylation sites from protein sequences using K-nearest neighbor algorithm via Chou's 5-step rule and pseudo components. J Theor Biol 470: 43-49.
- Salman, Khan M, Iqbal N, Hussain T, AfzalA S, et al. (2019) two-level computation model based on deep learning algorithm for identification of piRNA and their functions via Chou's 5-steps rule. International Journal of Peptide Research and Therapeutics (IJPRT).
- Tahir M, Tayara H, Chong KT (2019) iDNA6mA (5-step rule): Identification of DNA N6-methyladenine sites in the rice genome by intelligent computational model via Chou's 5-step rule. CHEMOLAB 189: 96-101.
- Vishnoi S, Garg P, Arora P (2019) Physicochemical n-Grams Tool: A tool for protein physicochemical descriptor generation via Chou's 5-steps rule. Chem Biol Drug Des.
- Wiktorowicz A, Wit A, Dziewierz A, Rzeszutko L, Dudek D, et al. (2019) Calcium Pattern Assessment in Patients with Severe Aortic Stenosis Via the Chou's 5-Steps Rule. Current Pharmaceutical Design 25(35): 3769-3775.
- Yang L, Lv Y, Wang S, Zhang Q, Y Pan, et al. (2019) Identifying FL11 subtype by characterizing tumor immune microenvironment in prostate adenocarcinoma via Chou's 5-steps rule. Genomics.
- Khan YD, Amin N, Hussain W, Rasool N, Khan SA, et al. (2020) iProtease-PseAAC(2L): A two-layer predictor for identifying proteases and their types using Chou's 5-step-rule and general PseAAC. Anal Biochem 588: 113477.
- Xu Y, Ding J, Wu LY, Chou KC (2013) iSNO-PseAAC: Predict cysteine S-nitrosylation sites in proteins by incorporating position specific amino acid propensity into pseudo amino acid composition PLoS ONE 8(2): e55844.
- Xu Y, Shao XJ, Deng NY, Chou KC (2013) iSNO-AAPair: incorporating amino acid pairwise coupling into PseAAC for predicting cysteine S-nitrosylation sites in proteins. PeerJ 1: e171.
- Xu Y, Wen X, Shao XJ, Deng NY, Chou KC (2014) iHyd-PseAAC: Predicting hydroxyproline and hydroxylysine in proteins by incorporating dipeptide position-specific propensity into pseudo amino acid composition. International Journal of Molecular Sciences (IJMS) 15: 7594-7610.
- Xu Y, Wen X, Wen LS, Wu LY, Deng NY, et al. (2014) iNitro-Tyr: Prediction of nitrotyrosine sites in proteins with general pseudo amino acid composition. PLoS ONE 9: e105018.
- Xu Y, Chou KC (2016) Recent progress in predicting posttranslational modification sites in proteins. Curr Top Med Chem 16: 591-603.
- Liu LM, Xu Y, Chou KC (2017) iPGK-PseAAC: identify lysine phosphoglycerylation sites in proteins by incorporating four different tiers of amino acid pairwise coupling information into the general PseAAC. Med Chem 13(6): 552-559.
- Xu Y, Li C, Chou KC (2017) iPreny-PseAAC: identify C-terminal cysteine prenylation sites in proteins by incorporating two tiers of sequence couplings into PseAAC. Med Chem 13: 544-551.
- Cai L, Wan CL, He L, Jong S, Chou KC (2015) Gestational influenza increases the risk of psychosis in adults. Medicinal Chemistry 11(7): 676-682.
- Liu J, Song J, Wang MY, He L, Cai L, et al. (2015) Association of EGF rs4444903 and XPD rs13181 polymorphisms with cutaneous melanoma in Caucasians. Medicinal Chemistry 11: 551-559.
- Cai L, Yuan W, Zhang Z, He L, Chou KC (2016) Modulation of cytokine network in the comorbidity of schizophrenia and tuberculosis. Curr Top Med Chem 16(6): 655-665.
- Cai L, Yuan W, Zhang Z, He L, Chou KC (2016) In-depth comparison of somatic point mutation callers based on different tumor next-generation sequencing depth data Scientific Reports 6: 36540.
- Zhu Y, Cong QW, Liu Y, Wan CL, Yu T, et al. (2017) Antithrombin is an importantly inhibitory role against blood clots. Curr Top Med Chem 16: 666-674.
- Zhang ZD, Liang K, Li K, Wang GQ, Zhang KW, et al. (2017) Chlorella vulgaris induces apoptosis of human non-small cell lung carcinoma (NSCLC) cells. Med Chem 13: 560-568.
- Cai L, Huang T, Su J, Zhang X, Chen W, et al. (2017) Implications of newly identified brain eQTL genes and their interactors in Schizophrenia. Molecular Therapy - Nucleic Acids 12: 433-442.
- Niu B, Zhang M, Du P, Jiang L, Qin R, et al. (2017) Small molecular floribundiquinone B derived from medicinal plants inhibits acetylcholinesterase activity. Oncotarget 8(34): 57149-57162.
- Su Q, Lu W, Du D, Chen F, Niu B, et al. (2017) Prediction of the aquatic toxicity of aromatic compounds to tetrahymena pyriformis through support vector regression. Oncotarget 8(30): 49359-49369.
- Lu Y, Wang S, Wang J, Zhou G, Zhang Q, et al. (2019) An Epidemic Avian Influenza Prediction Model Based on Google Trends. Letters in Organic Chemistry 16: 303-310.
- Niu B, Liang C, Lu Y, Zhao M, Chen Q, et al. (2019) Glioma stages prediction based on machine learning algorithm combined with protein-protein interaction networks. Genomics.
- Jia J, Liu Z, Xiao X, Liu B, Chou KC (2016) Identification of protein-protein binding sites by incorporating the physicochemical properties and stationary wavelet transforms into pseudo amino acid composition (iPPBS-PseAAC). J Biomol Struct Dyn (JBSD) 34: 1946-1961.
- Jia J, Liu Z, Xiao X, Liu B, Chou KC (2016) iSuc-PseOpt: Identifying lysine succinylation sites in proteins by incorporating sequence-coupling effects into pseudo components and optimizing imbalanced training dataset. Anal Biochem 497: 48-56.
- Jia J, Liu Z, Xiao X, Liu B, Chou KC (2016) pSuc-Lys: Predict lysine succinylation sites in proteins with PseAAC and ensemble random forest approach. Journal of Theoretical Biology 394: 223-230.
- Jia J, Liu Z, Xiao X, Liu B, Chou KC (2016) iCar-PseCp: identify carbonylation sites in proteins by Monto Carlo sampling and incorporating sequence coupled effects into general PseAAC. Oncotarget 7(23): 34558-34570.
- Jia J, Liu Z, Xiao X, Liu B, Chou KC (2016) iPPBS-Opt: A Sequence-Based Ensemble Classifier for Identifying Protein-Protein Binding Sites by Optimizing Imbalanced Training Datasets. Molecules 21(1): E95.
- Jia J, Zhang L, Liu Z, Xiao X, Chou KC (2016) pSumo-CD: Predicting sumoylation sites in proteins with covariance discriminant algorithm by incorporating sequence-coupled effects into general PseAAC. Bioinformatics 32(20): 3133-3141.
- Liu Z, Xiao X, Yu DJ, Jia J, Qiu WR, et al. (2016) pRNAm-PC: Predicting N-methyladenosine sites in RNA sequences via physical-chemical properties. Anal Biochem 497: 60-67.
- Xiao X, Ye HX, Liu Z, Jia JH, Chou KC (2016) iROS-gPseKNC: predicting replication origin sites in DNA by incorporating dinucleotide position-specific propensity into general pseudo nucleotide composition. Oncotarget 7(23): 34180-34189.
- Qiu WR, Sun BQ, Xiao X, Xu ZC, Jia JH, et al. (2018) iKcr-PseEns: Identify lysine crotonylation sites in histone proteins with pseudo components and ensemble classifier. Genomics 110(5): 239-246.
- Jia J, Li X, Qiu W, Xiao X, Chou KC (2019) iPPI-PseAAC(CGR): Identify protein-protein interactions by incorporating chaos game representation into PseAAC. Journal of Theoretical Biology 460: 195-203.
- Chen W, Ding H, Feng P, Lin H, Chou KC (2016) iACP: a sequence-based tool for identifying anticancer peptides. Oncotarget 7(13): 16895-16909.
- Chen W, Feng P, Yang H, Ding H, Lin H, et al. (2016) Using deformation energy to analyze nucleosome positioning in genomes. Genomics 107(2-3): 69-75.
- Chen W, Tang H, Ye J, Lin H, Chou KC (2016) iRNA-PseU: Identifying RNA pseudouridine sites Molecular Therapy - Nucleic Acids 5: e332.
- Zhang CJ, Tang H, Li WC, Lin H, Chen W, et al. (2016) iOri-Human: identify human origin of replication by incorporating dinucleotide physicochemical properties into pseudo nucleotide composition. Oncotarget 7(43): 69783-69793.
- Chen W, Feng P, Yang H, Ding H, Lin H, et al. (2017) iRNA-AI: identifying the adenosine to inosine editing sites in RNA sequences. Oncotarget 8(3): 4208-4217.
- Feng P, Ding H, Yang H, Chen W, Lin H, et al. (2017) iRNA-PseColl: Identifying the occurrence sites of different RNA modifications by incorporating collective effects of nucleotides into PseKNC. Molecular Therapy - Nucleic Acids 7: 155-163.
- Chen W, Ding H, Zhou X, Lin H, Chou KC (2018) iRNA(m6A)-PseDNC: Identifying N6-methyladenosine sites using pseudo dinucleotide composition. Analytical Biochemistry 561-562: 59-65.
- Chen W, Feng P, Ding H, Lin H, Chou KC (2018) iRNA-3typeA: identifying 3-types of modification at RNA’s adenosine sites. Molecular Therapy: Nucleic Acid 11: 468-474.
- Su ZD, Huang Y, Zhang ZY, Zhao YW, Wang D, et al. (2018) iLoc-lncRNA: predict the subcellular location of lncRNAs by incorporating octamer composition into general PseKNC. Bioinformatics 34(24): 4196-4204.
- Yang H, Qiu WR, Liu G, Guo FB, Chen W, et al. (2018) iRSpot-Pse6NC: Identifying recombination spots in Saccharomyces cerevisiae by incorporating hexamer composition into general PseKNC International Journal of Biological Sciences 14(8): 883-891.
- Feng P, Ding H, Lin H, Chen W, Chou KC (2017) iDNA6mA-PseKNC: Identifying DNA N(6)-methyladenosine sites by incorporating nucleotide physicochemical properties into PseKNC. Genomics 111(1): 96-102.
- Du QS, Wang SQ, Wang QY, Huang RB, Chou KC (2017) 2L-PCA: A two-level principal component analyzer for quantitative drug design and its applications. Oncotarget 8(41): 70564-70578.
- Liu B, Fang L, Long R, Lan X, Chou KC (2017) iEnhancer-2L: a two-layer predictor for identifying enhancers and their strength by pseudo k-tuple nucleotide composition. Bioinformatics 32(3): 362-369.
- Liu B, Long R, Chou KC (2016) iDHS-EL: Identifying DNase I hypersensi-tivesites by fusing three different modes of pseudo nucleotide composition into an ensemble learning framework. Bioinformatics 32(16): 2411-2418.
- Liu B, Wang S, Long R, Chou KC (2017) iRSpot-EL: identify recombination spots with an ensemble learning approach. Bioinformatics 33(1): 35-41.
- Liu B, Yang F, Chou KC (2017) 2L-piRNA: A two-layer ensemble classifier for identifying piwi-interacting RNAs and their function. Molecular Therapy - Nucleic Acids 7: 267-277.
- Liu B, Li K, Huang DS, Chou KC (2018) iEnhancer-EL: Identifying enhancers and their strength with ensemble learning approach. Bioinformatics 34(22): 3835-3842.
- Liu B, Weng F, Huang DS, Chou KC (2018) iRO-3wPseKNC: Identify DNA replication origins by three-window-based PseKNC. Bioinformatics 34(18): 3086-3093.
- Qiu WR, Sun BQ, Xiao X, Xu D, Chou KC (2018) iPromoter-2L: a two-layer predictor for identifying promoters and their types by multi-window-based PseKNC. Bioinformatics 34(1): 33-40.
- Qiu WR, Sun BQ, Xiao X, Xu D, Chou KC (2016) iHyd-PseCp: Identify hydroxyproline and hydroxylysine in proteins by incorporating sequence-coupled effects into general PseAAC. Oncotarget 7(28): 44310-44321.
- Qiu WR, Sun BQ, Xiao X, Xu D, Chou KC (2016) iPTM-mLys: identifying multiple lysine PTM sites and their different types. Bioinformatics 32(20): 3116-3123.
- Qiu WR, Xiao X, Chou KC (2017) iPhos-PseEn: identifying phosphorylation sites in proteins by fusing different pseudo components into an ensemble classifier. Oncotarget 7(32): 51270-51283.
- Qiu WR, Jiang SY, Sun BQ, Xiao X, Cheng X, et al. (2017) iRNA-2methyl: identify RNA 2′-O-methylation sites by incorporating sequence-coupled effects into general PseKNC and ensemble classifier. Medicinal Chemistry 13(8): 734-743.
- Qiu WR, Jiang SY, Xu ZC, Xiao X, Chou KC (2017) RNAm5C-PseDNC: identifying RNA 5-methylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition. Oncotarget 8(25): 41178-41188.
- Qiu WR, Sun BQ, Xiao X, Xu D, Chou KC (2017) iPhos-PseEvo: Identifying human phosphorylated proteins by incorporating evolutionary information into general PseAAC via grey system theory. Molecular Informatics 36 UNSP 1600010.
- Zhai X, Chen M, Lu W (2018) Accelerated search for perovskite materials with higher Curie temperature based on the machine learning methods. Computational Materials Science 151: 41-48.
- Chou KC (2011) Some remarks on protein attribute prediction and pseudo amino acid composition (50th Anniversary Year Review, 5-steps rule). Journal of Theoretical Biology 273: 236-247.
- Chou KC (2019) Impacts of pseudo amino acid components and 5-steps rule to proteomics and proteome analysis. Current Topics in Medicinak Chemistry (CTMC) (Special Issue ed. G.P Zhou) 19(25): 2283-2300.
- Chou KC (2019) Two kinds of metrics for computational biology. Genomics.
- Chou KC (2019) Proposing pseudo amino acid components is an important milestone for proteome and genome analyses. International Journal for Peptide Research and Therapeutics (IJPRT).
- Chou KC (2019) An insightful recollection for predicting protein subcellular locations in multi-label systems. Genomics.
- Chou KC (2019) Progresses in predicting post-translational modification. International Journal of Peptide Research and Therapeutics (IJPRT).
- Chou KC (2019) Recent Progresses in Predicting Protein Subcellular Localization with Artificial Intelligence (AI) Tools Developed Via the 5-Steps Rule. Japanese Journal of Gastroenterology and Hepatology 2: 1-4.
- Chou KC (2019) An insightful recollection since the distorted key theory was born about 23 years ago. Genomics.
- Chou KC (2019) Artificial intelligence (AI) tools constructed via the 5-steps rule for predicting post-translational modifications. Trends in Artificial Intelligence (TIA) 3: 60-74.
- Chou KC (2019) Distorted Key Theory and Its Implication for Drug Development. Current Genomics.
- Chou KC (2019) An Insightful 10-year Recollection Since the Emergence of the 5-steps Rule. Current Pharmaceutical Design 25: 4223-4234.
- Chou KC (2009) An insightful recollection since the birth of Gordon Life Science Institute about 17 years ago. Advancement in Scientific and Engineering Research.
- http://doi.org/10.33495/aser_v4i2.19.105.
- Chou KC (2019) Gordon Life Science Institute: Its philosophy, achievements, and perspective. Annals of Cancer Therapy and Pharmacology.

 Short Communication
Short Communication