Abstract
Receptor-interacting serine/threonine protein kinase 1 (RIPK1) is able to decide the fate of a cell in response to different stimuli, activating pathways leading to cell survival, cell death and inflammation. Necroptosis, a new and well documented type of programmed cell death, is involved in several human diseases including cancer. Further, the fact that programmed cell death can be molecularly controlled has prompted increasing scientific interest in the action of several so-called “epi-drugs”. Starting from necrostatin-1, many small molecules inhibiting RIPK1 have been characterized. Although some are currently in use in the clinic for inflammatory diseases, no robust evidence of their anticancer effect has been reported to date. The recently characterized sirtuin inhibitor MC2494 modulates RIPK1 function through its acetylation and is able to selectively activate cell death pathways in cancer. Here, we provide a greater insight into the molecular mechanism of action of MC2494, highlighting its ability to directly target RIPK1 within the molecular complex activated. Our findings support the key role of MC2494 in regulating RIPK1-mediated cell death, making necroptosis a pharmacologically druggable event useful in cancer therapy.
Keywords: Necroptosis; Cancer; RIPK1; SIRT inhibitor
Introduction
Programmed cell death (PCD) is a fundamental process involved in animal development and in maintaining cellular homeostasis [1,2]. In addition to the canonical pathways of cell death, a new form of programmed necrosis called “necroptosis” has recently emerged [2]. Necroptosis displays features of both apoptosis and necrosis, the major models of cellular death, and is characterized by cytosolic shrinkage and DNA fragmentation, which eventually lead to inflammation via the secretion of cytokines and chemokines [3]. Necroptosis is mediated by the interaction of receptorinteracting serine/threonine protein kinase (RIPK) 1 and 3 when caspase-8 is not active [4]. RIPK1 has a functional role, interpreting and transmitting extracellular signals through the activation of downstream pathways by different molecular interactors, including RIPK3 and mixed- lineage kinase domain-like pseudokinase (MLKL) [5,6]. RIPK1 is known to be an important upstream kinase in several pathways able to regulate inflammation [7], also through direct regulation of necroptosis [8,9]. Starting from the identification and characterization of necrostatins (Nec) [10,11], other RIPK1 inhibitors have been characterized [7,12,13]. Although these inhibitors were shown to act as promising agents for the treatment of inflammatory diseases in clinical trials [14], their use in cancer therapy has yet to be well described. A recent study illustrated the action of GNE684, a novel RIPK1 inhibitor, which binds the same hydrophobic pocket in the RIPK1 kinase domain as necrostatins [15]. GNE684 can block inflammatory processes but not tumour growth. Another very recent report identified a new RIPK1 inhibitor that promotes a tumour-suppressive T cell phenotype in pancreatic adenocarcinoma, highlighting a previously unknown role for RIPK1 in cancer [12]. RIPK1 and RIPK3 are found deregulated in many cancer types and are often associated with poor outcomes [16-20]. Reducing uncontrolled cell proliferation by targeting both these kinases may thus represent a novel anticancer strategy. Since cancer is also considered an epigenetic disease, the identification of new compounds that modify DNA and chromatin structure (known as “epi-drugs”) and are therefore able to reprogram cell fate is of great clinical importance [21,22]. Sirtuins (SIRTs), class III NAD+- dependent histone deacetylases, are known to play an important biological role in regulating many cellular processes involved in cancer initiation and progression such as DNA repair, metabolism, cell proliferation and cell death [23,24]. The functional involvement of SIRTs in cancer and their already reported role as either tumour suppressors or tumour promoters has led to increased scientific interest in this enzyme family [25-27].
A new SIRT inhibitor (SIRTi), MC2494, a derivative of the wellknown SIRT2i AGK2 [28,29], was recently characterized [30]. This promising compound is able to block cancer growth by activating RIPK1-mediated necroptotic death pathway in a tumour-selective manner [30]. The concomitant presence of RIPK1 in molecular complexes of proteins exerting different enzymatic activities, such as deacetylases, acetyltransferases and kinases, supports the idea that interaction between proteins is involved in the activation of many important biological pathways. The documented cell death activity of MC2494 prompted us to further explore the RIPK1- mediated mechanism of cell death activated by MC2494 and its role as a potential necroptosis inhibitor.
Materials and Methods
Cell Line, Ligands and Antibodies
Cell line: U937 cell were purchased from DSMZ and grown following standard protocols at 37 °C with 5% CO2 in Roswell Park Memorial Institute (RPMI) 1640 medium, with 10% fetal bovine serum (Gibco), 2 mM L-glutamine (Euroclone) and antibiotics (100 U/mL penicillin, 100 μg/mL streptomycin and 250 ng/mL amphotericin-B; Euroclone). The cell line was tested and authenticated, and mycoplasma contamination was regularly examined using EZ-PCR Mycoplasma Test Kit (Biological Industries). Cells were used for experiments between passages 10 to 20 and then discarded. Ligands: MC2494 was prepared as previously reported [30], Nec-1 (Sigma) was used at 50 μM. Antibodies: RIPK1 was purchased from BD Biosciences, RIPK3 and Actin were purchased from Santa Cruz, MLKL and GAPDH were purchased from Cell Signaling.
Cell Death Analysis
Cells were plated (2 × 105 cells/mL) and after stimulation were harvested with PBS, centrifuged at 1200 rpm for 5 min, and resuspended in i) 500 μL of a hypotonic solution containing 1X PBS, 0.1% sodium citrate, 0.1% NP-40, RNAase A and 50 mg/mL PI (for sub-G1 evaluation), and in ii) 500 μL 1X PBS and 0.2 mg/ mL propidium iodide (PI; for PI evaluation). The results were acquired on a FACS Calibur (BD Biosciences) and a BD Accuri TM C6 flow cytometer system (BD Biosciences). Each experiment was performed in biological triplicates and values expressed as mean ± standard deviation.
Annexin V/PI Double Staining
Annexin V evaluation was performed as suggested by the supplier (Dojindo). Briefly, cells were suspended in Annexin V binding solution at the concentration of 1 × 106 cells/mL Five μL of fluorescein isothiocyanate- conjugated Annexin V and 5 μL PI were added to 100 μL cellular suspension transferred into a new tube. Reaction was carried out for 15 min at room temperature. The results were acquired on a FACS Calibur (BD Biosciences. Graphs show the experimental results of biological triplicates.
Statistical Analysis
Graphs shown are the results of three independent experiments with an error bar indicating standard deviation. Differences between treated cells versus control cells were analyzed using GraphPad Prism 6.0 software (GraphPad Software). Statistical comparison was performed by applying one-way analysis of variance (ANOVA) and Dunnett’s multiple-comparison test. Differences between groups were significant at a p- value < 0.05.
Protein Extraction
Cell pellets were washed in PBS and suspended in lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride and protease inhibitor. The lysis reaction was carried out for 15 mins at 4 °C and centrifuged at 13,000 rpm for 30 min at 4 °C. Protein concentration was quantified by Bradford assay (Bio-Rad).
Western Blot
Fifty μg of proteins was loaded onto 10–15% polyacrylamide gels and transferred to nitrocellulose membrane using a transfer apparatus according to the manufacturer’s protocols (Bio-Rad). Membrane was blocked with 5% non-fat milk in TBST (10 mM Tris pH 8.0, 150 mM NaCl, 0.5% TWEEN 20) for 60 min, the membrane was washed once with TBST and incubated with antibodies. Detection was performed with an Enhanced chemiluminescence (ECL) system (Amersham Biosciences) according to the manufacturer’s protocol. Western blots were normalized through densitometry analysis, performed using the Image J Gel Analysis tool.
Living Cell Image Detection
U937 cells were plated in 48 wells 2 × 105 cells/mL and induced with MC2494 at 50 μM. After 24 h, the cells were treated with Hoechst Stain solution (B-Bridge) to evaluate the living cells, and were analyzed by Cytation 5 Cell Imaging Multi-Mode Reader (BioTeK).
Chemical Modelling
The RIP1 kinase active monomer in complex with GNE684 (PDB: 6NYH) was used for docking purposes. We used the DundeePRODRG2 Server to build the energy-minimized 3D-structure of MC2494. Docking of MC2494 was carried out using Molegro Virtual Docker (CLCbio). The flexible torsions of MC2494 were automatically assigned and checked for consistency. The 3D-structure of human RIP1 was automatically prepared by adding hydrogens, charges and Tripos atom type potentials. During docking, a search space of 10Å radius in the active site cavity was used. MolDock score was used as scoring function and Simplest Evolution was used as search algorithm. Ten runs were defined. Other parameters were set at their default. Similar docking solutions (RMSD 1.0 Å) were clustered, and the best one was selected as representative.
Results and Discussion
Study of Cell Death Induced by MC2494
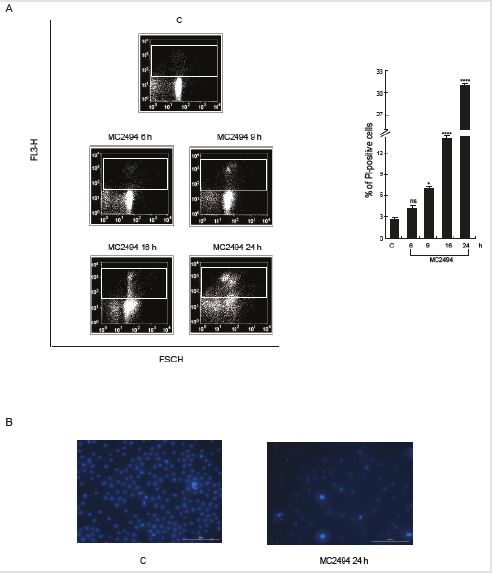
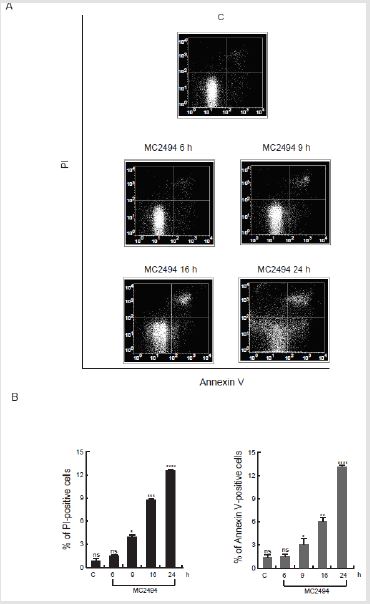
We previously reported that treatment with MC2494 determines a strong increase in cell death in many cancer cell lines [30,31]. To better investigate MC2494-mediated induction of cell death, a time-course experiment was performed in U937 leukaemia cells and the percentage of cells positive to PI was measured. As expected, MC2494 determined a reduction in cell viability, showing a slight effect after 9 h of induction (7.74% of PI- positive cells) and a greater increase at the longer treatment times of 16 h and 24 h (14.4% and 30.8% of PI- positive cells, respectively) (Figure 1A). Cell death was confirmed by Hoechst staining. After 24 h of treatment, many apoptotic features such as small- sized cells, reduction in proliferation, and small and condensed nuclei became more apparent (Figure 1B). Since PI is a marker used to assess the integrity of plasmatic and nuclear membranes lost as a result of necrosis or late stage events of apoptosis, in order to extend the study of cell death and better characterize the apoptotic pathway involved, Annexin V/PI assays were performed. Double staining was carried out in U937 cells treated with MC2494 at 50 μM for different times of induction (6–24 h). Early and late stage apoptotic cells were detected and analysed by fluorescence-activated cell sorting (FACS). After 16 h and 24 h of induction, an increase in apoptotic population at early stage (5.39% and 12.40%, respectively) and at late stage (8.99% and 12.13%, respectively) was observed (Figure 2).
Figure 1: Cell death evaluation.
A. FACS analysis of membrane permeabilization by PI. PI staining was performed in U937cells with the compounds at 50
μM concentration at indicated times of induction. Left panel: FACS density plot; Right panel: PI analysis.
B. Hoechst coloration. Objective lenses 20X. Graphs show the mean of at least three independent experiments with error
bars indicating standard deviation. Values are mean ± SD of biological triplicates.
****p-value ≤ 0.0001, ***p-value ≤ 0.001, **p-value ≤ 0.01, *p-value ≤ 0.05, ns p-value > 0.05 vs. control cel
Figure 2: Study of apoptosis event. Annexin V/PI assays were performed in U937 cells with MC2494 at 50 μM concentration at indicated times of induction.
A. Density plots of Annexin V/PI evaluation.
B. Annexin V/PI analysis; Left panel: PI-positive cells; Right panel: Annexin V-positive cells. Graphs show the mean of
three independent experiments with error bars indicating standard deviation. Values are mean ± SD of biological triplicates.
****p-value ≤ 0.0001, ***p-value ≤ 0.001, **p-value ≤ 0.01, *p-value ≤ 0.05, ns p-value > 0.05 vs. control cells.
MC2494 Modulates RIPK1
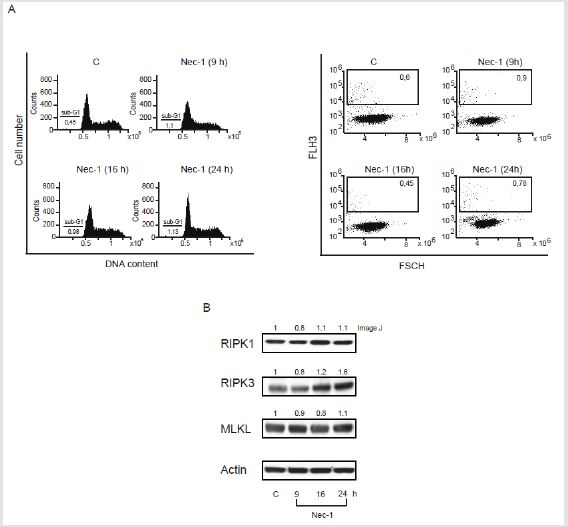
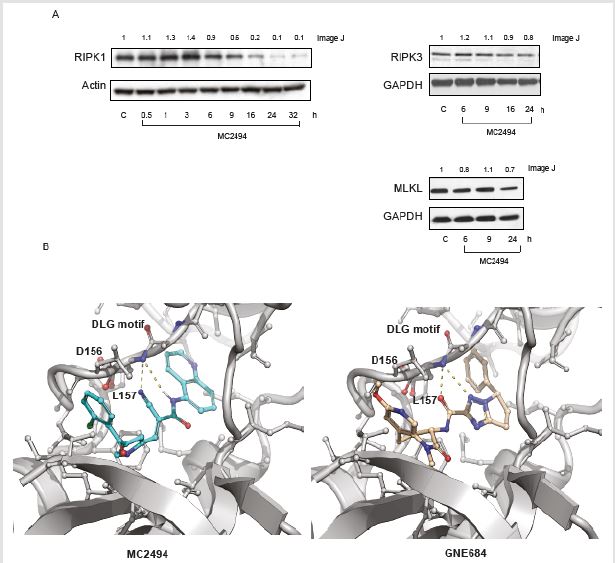
FACS analysis performed in U937 cells showed that Nec-1 treatment at 50 μM for different times (924 h ) was not able to induce cell death, as demonstrated by the percentage of cells in sub-G1 phase as well as positivity to PI (Figure 3A). Significantly, Nec-1 did not affect necrosome signalling pathway as shown by RIPK1, RIPK3 and MLKL protein levels (Figure 3B). Our previous study indicated that MC2494 regulates necroptotic pathway via RIPK1 protein regulation. To gain further mechanistic insights, RIPK1 expression was evaluated by Western blot analysis in U937 cells treated with MC2494 at 50 μM concentration for different times (0.532 h). As expected, the longer times of induction at 24 h and 32 h resulted in a strong decrease in protein expression, which was already evident after 9 h of treatment when a 40% decrease in RIPK1 protein expression was observed (Figure 4A). Based on this observation, the expression of necrosome formation was investigated by studying RIPK3 and MLKL protein expression. No substantial modulation was observed for RIPK3 and MLKL after MC2494 treatment (Figure 4A). Taken together these results indicate that, unlike Nec-1, MC2494 is able to determine a decrease in RIPK1 protein expression.
Figure 3: Nec-1 cell death and necrosome evaluation.
A. Sub-G1 (Left panel) and PI (Right panel) evaluations performed in U937 cells with Nec-1 at 50 M concentration at
indicated times of induction.
B. Western blot analysis of RIPK1, RIPK3 and MLKL. Actin was used as loading control. Numbers on Western blot indicate
the result of densitometry analysis, performed using the Image J Gel Analysis tool.
Figure 4: MC2494 regulates necrosome.
A. Western blot analysis of RIPK1, RIPK3 and MLKL. Actin and GAPDH were used as loading controls.
B. Cartoon representation of the docked pose of MC2494 (cyan sticks – Left Panel) and B) GNE684 (pink sticks – Right
Panel) into the experimentally determined complex with RIP1 (PDB: 6NYH). Nitrogen and oxygen atoms are colored blue
and red, respectively. Polar interactions between the compounds and the protein moiety are shown in yellow dashed lines.
Residues are labelled according to sequence numbering. Numbers on Western blot indicate the result of densitometry analysis,
performed using the Image J Gel Analysis tool.
Modelling of MC2494 into the Active Site of RIP1
A docked model of MC2494 showed that the compound binds to the same hydrophobic pocket as and in a similar conformation to GNE684, a newly characterized RIP1K inhibitor (Figure 4B). In this model, MC2494 binds to an inactive conformation of RIPK1, similar to Nec-1, GNE684 and other type II kinase inhibitors, with Asp156 and Leu157 of the DLG motif (commonly DFG in other kinases) in the “out” conformation. The computed interaction energy as assessed by MVD scoring function (-132.7) is comparable to that measured for GNE684 (-148.4), suggesting that MC2494 may act as a RIPK1 inhibitor with comparable efficacy to GNE684.
Conclusion
The mechanism of action of MC2494 is fully in line with the concept of epigenetic therapy, which aims to restore the correct execution of altered and silenced cellular pathways. The observed RIPK1-mediated cell death induced by MC2494 already at the early treatment time strengthens its potential role as an anticancer agent in leukaemia. Further molecular and enzymatic investigations will be necessary to assess the ability of MC2494 to modulate RIP1 kinase activity. This work provides the basis for a future structurebased drug design study of small molecules able to modulate RIPK1, and thereby induce cancer-selective cell death.
Author Contributions
Funding
This work was supported by the VALERE: Vanvitelli per la Ricerca Program, AIRC – 20447- 17217 and the Italian Ministry of Health (GR-2018-12366268); MIUR20152TE5PK; Campania Regional Government Technology Platform Lotta alle Patologie Oncologiche: iCURE; Campania Regional Government FASE2: IDEAL. MIUR, Proof of Concept POC01_00043.
Acknowledgments
We thank C. Fisher for linguistic editing.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Fuchs Y, Steller H (2011) Programmed cell death in animal development and disease. Cell 147(4): 742-758.
- Zhou W, Yuan J (2014) Necroptosis in health and diseases. Seminars in cell & developmental biology 35: 14-23.
- Choi ME, Price DR, Ryter SW, Choi AMK (2019) Necroptosis: a crucial pathogenic mediator of human disease. JCI insight 4(15): 128834.
- Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P (2007) RIP1, a kinase on the crossroads of a cell's decision to live or die. Cell death and differentiation 14(3): 400-410.
- Dhuriya YK, Sharma D (2018) Necroptosis: a regulated inflammatory mode of cell death. Journal of neuroinflammation 15(1): 199.
- Vanden Berghe T, Kaiser WJ, Bertrand MJ, Vandenabeele P (2015) Molecular crosstalk between apoptosis, necroptosis, and survival signaling. Molecular & cellular oncology 2(4): e975093.
- Harris PA, Berger SB, Jeong JU, Nagilla R, Bandyopadhyay D, et al. (2017) Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases. Journal of medicinal chemistry 60(4): 1247-1261.
- Christofferson DE, Li Y, Hitomi J, Zhou W, Upperman C, et al. (2012) A novel role for RIP1 kinase in mediating TNFalpha production. Cell death & disease 3: e320.
- Duong BH, Onizawa M, Oses Prieto JA, Advincula R, Burlingame A, et al. (2015) A20 restricts ubiquitination of pro-interleukin-1beta protein complexes and suppresses NLRP3 inflammasome activity. Immunity 42(1): 55-67.
- Degterev A, Hitomi J, Germscheid M, Chen IL, Korkina O, et al. (2008) Identification of RIP1 kinase as a specific cellular target of necrostatins. Nature chemical biology 4(5): 313-321.
- Xie T, Peng W, Liu Y, Yan C, Maki J, et al. (2013) Structural basis of RIP1 inhibition by necrostatins. Structure 21(3): 493-499.
- Harris PA, Marinis JM, Lich JD, Berger SB, Chirala A, et al. (2019) Identification of a RIP1 Kinase Inhibitor Clinical Candidate (GSK3145095) for the Treatment of Pancreatic Cancer. ACS medicinal chemistry letters 10(6): 857-862.
- Zhou T, Wang Q, Phan N, Ren J, Yang H, et al. (2019) Identification of a novel class of RIP1/RIP3 dual inhibitors that impede cell death and inflammation in mouse abdominal aortic aneurysm models. Cell death & disease 10(3): 226.
- Lukens JR, Vogel P, Johnson GR, Kelliher MA, Iwakura Y, et al. (2013) RIP1-driven autoinflammation targets IL-1alpha independently of inflammasomes and RIP3. Nature 498(7453): 224-227.
- Patel S, Webster JD, Varfolomeev E, Kwon YC, Cheng JH, et al. (2019) RIP1 inhibition blocks inflammatory diseases but not tumor growth or metastases. Cell death and differentiation.
- Degterev A, Ofengeim D, Yuan J (2019) Targeting RIPK1 for the treatment of human diseases. Proceedings of the National Academy of Sciences of the United States of America 116(20): 9714-9722.
- Gong Y, Fan Z, Luo G, Yang C, Huang Q, et al. (2019) The role of necroptosis in cancer biology and therapy. Molecular cancer 18(1): 100.
- Hanggi K, Vasilikos L, Valls AF, Yerbes R, Knop J, et al. (2017) RIPK1/RIPK3 promotes vascular permeability to allow tumor cell extravasation independent of its necroptotic function. Cell death & disease 8(2): e2588.
- Moriwaki K, Chan FK (2013) RIP3: A molecular switch for necrosis and inflammation. Genes & development 27(15): 1640-1649.
- Schneider AT, Gautheron J, Feoktistova M, Roderburg C, Loosen SH, et al. (2017) RIPK1 Suppresses a TRAF2- Dependent Pathway to Liver Cancer. Cancer cell 31(1): 94-109.
- Miranda Furtado CL, Dos Santos Luciano MC, Silva Santos RD, Furtado GP, Moraes MO, et al. (2019) Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics 14(12): 1164-1176.
- Nebbioso A, Tambaro FP, Dell Aversana C, Altucci L (2018) Cancer epigenetics: Moving forward. PLoS genetics 14(6): e1007362.
- Lin Z, Fang D (2013) The Roles of SIRT1 in Cancer. Genes & cancer 4(3-4): 97-104.
- Vassilopoulos A, Fritz KS, Petersen DR, Gius D (2011) The human sirtuin family: evolutionary divergences and functions. Human genomics 5(5): 485-496.
- Bosch Presegue L, Vaquero A (2011) The dual role of sirtuins in cancer. Genes & cancer 2(6): 648-662.
- Carafa V, Altucci L, Nebbioso A (2019) Dual Tumor Suppressor and Tumor Promoter Action of Sirtuins in Determining Malignant Phenotype. Frontiers in pharmacology 10: 38.
- O Callaghan C, Vassilopoulos A (2017) Sirtuins at the crossroads of stemness, aging, and cancer. Aging cell 16(6): 1208-1218.
- Kim HW, Kim SA, Ahn SG (2016) Sirtuin inhibitors, EX527 and AGK2, suppress cell migration by inhibiting HSF1 protein stability. Oncology reports 35(1): 235-242.
- Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, et al. (2007) Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science 317(5837): 516-519.
- Carafa V, Nebbioso A, Cuomo F, Rotili D, Cobellis G, et al. (2018) RIP1-HAT1-SIRT Complex Identification and Targeting in Treatment and Prevention of Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research 24(12): 2886-2900.
- Carafa V, Poziello A, Della Torre L, Giovannelli P, Di Donato M, et al. (2019) Enzymatic and Biological Characterization of Novel Sirtuin Modulators against Cancer. International journal of molecular sciences 20(22).

 Research Article
Research Article