Abstract
Purpose: Inflammation is well known to be a key culprit behind tumor promotion and metastasis in Triple Negative Breast Cancer. In hopes of further understanding TNBC pro-inflammatory drug effects, we have studied the current first-line therapy drug doxorubicin along with a second-generation proteasome inhibitor that demonstrate a reduction in inflammation. In this study, we focused on conducting a gene array to elucidate the effects of carfilzomib and doxorubicin on pro-inflammatory gene expression in both MCF-10A and MDA-MB-231 cells.
Methods: 93 Taqman Human Inflammatory Gene Microarray design. MCF-10A normal breast cells and MDA-MB-231 TNBC cells were treated or left untreated with vehicle control (0.1% DMSO), Carfilzomib (5 nM), Doxorubicin (30 nM), or a combination of both. Control cDNA of the housekeeping gene 18S was used for signal normalization and expression changes were assessed using qt-PCR.
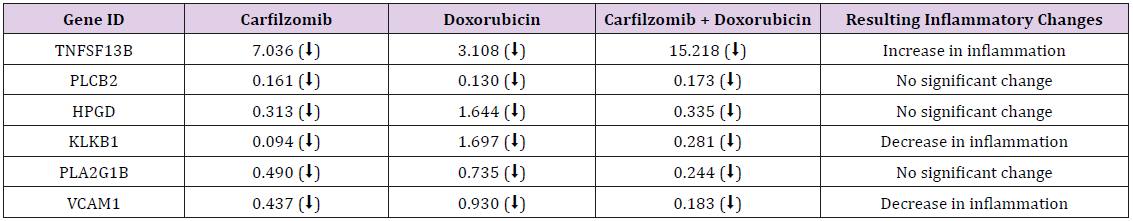
Results: Genes expression fold changes were analyzed and a comparison between MCF-10A and MDA-MB-231 cell lines was made. The design resulted in 6 main genes being identified as significant inflammatory modulators: HPGD, KLKB1, TNFSF13B, VCAM1, IL1RAPL2, and PTAFR.
Conclusion: Both carfilzomib and doxorubicin are seen to suppress several upregulated pro-inflammatory genes in MDA-MB-231 cells. Most notably, KLKB1, IL1RAPL2 and VCAM1 pro-inflammatory genes demonstrated drug-induced inflammatory suppression. On the other hand, HPGD, TNFSF13B, and PTAFR genes showed proinflammation upon giving the specific drug regimen. This suggests that these genes, along with the others identified through microarray, are significantly might be involved in inflammatory pathways. Further studies are needed to assess how their down regulation these gene expression changes may serve to supplement TNBC therapy.
Keywords: Doxorubicin; Cancer; Gene-Array; Inflammation; Proteasome Inhibitors; TNBC
Introduction
Approximately 15-20% of breast cancer cases are diagnosed as Triple Negative Breast Cancer (TNBC), where there is a lack of expression of the Estrogen Receptor (ER), Progesterone Receptor (PR), and epidermal growth factor receptor 2 (HER2). This subgroup unfortunately has the worst prognosis with relapse and metastasis increasingly common in patients. A significant challenge in cancer therapy and drug efficacy is overcoming the tumor promoting effects of inflammation, induced by both therapeutic drugs as well as the malignancy itself. Effects on a cellular level include increased proliferation, survival of malignant cells, and promotion of angiogenesis and metastasis Kanaya N [1]; Mantovani et al. [2]. The resultant secretion of chemokines leads to increased cell motility, invasiveness, and survival in response to inflammation Mantovani et al. [2]. This is especially prominent in TNBC.
Several first line chemotherapeutic drugs target TNBC, along with many other cancers, -Doxorubicin being one that is more commonly used. Studies have demonstrated that doxorubicin, although potent, further induces inflammatory processes Lopez- Hisijos N [3]. Doxorubicin also exhibits high short and long-term morbidity and mortality with severe heart failure occurring in more than 20% of patients Stagg and Allard [4]. In an effort to reduce these toxic effects, there is a lifetime dose limit; however, this commonly leads to under treatment of patients and eventual recurrence. This drug increases production of Transforming Growth Factor Beta (TGFB), leading to epithelial-mesenchymal transition and subsequent cell invasion in human MDA-MB-231 cell lines Bandyopadhyay et al. [5]; Vyas D [6]. Thus, a better understanding of the role of inflammation in TNBC treatment is imperative.
Carfilzomib (CARF) is a second-generation epoxiketone-based proteasome inhibitor that binds irreversibly to the 20S proteolytic core of the 26S proteasome Riz et al. [7]. Proteasome inhibitors are known to trigger anti-inflammatory effects within various cell lineages. The ability of CARF to not only kill tumor cells but also block the inflammatory pathways characteristic of TNBC suggests that CARF might have an effective therapeutic role in this disease. By utilizing CARF alongside doxorubicin, we are able to understand on a molecular level their role in TNBC, and further explore their mode of action on gene regulation.
In this current study, we focused on identifying the effects of carfilzomib, doxorubicin, or a combination of both on proinflammatory gene expression in MDA-MB-231 cells. Using quantitative PCR assay (TaqMan Human Inflammatory Assay) containing 93 genes and 3 control genes, we investigated whether expression of any of these genes may be up-regulated in the MDAMB- 231 cell line. By assessing specific inflammatory genes that are critical for the tumorogenicity of MDA-MB-231 cells, we endeavored to gain insight into the effect of the experimental drugs and their potential for offering an enhanced form of tumor suppression.
Methods
MCF-10A (normal mammary breast tissue) cells were obtained from the laboratory of Dr. Fluck, Michigan State University and MDA-MB-231 (triple negative breast cancer) cells were obtained from Dr. Gallo’s laboratory, Michigan State University. MCF-10A Cells were grown in MEBM growth medium and MDA-MB-231 cells were grown in Leibovitz’s L-15 growth medium. Cells (1X105) were plated in complete growth medium in six-well plates for overnight. Shortly after, the MCF-10A and MDA-MB-231 cells were treated or left untreated cells with vehicle control (0.1% DMSO), Carfilzomib (5 nM), Doxorubicin (30 nM), or a combination of both in complete growth medium for 24hrs. The concentrations of Carfilzomib and Doxorubicin used were in accordance with previous studies conducted in the lab Vyas et al. [8]. Total RNA was extracted using RNeasy Mini Kit (Qiagen, Catalog # 74104). The RNA conc. was measured using Nanodrop and RNA quality was checked using an Agilent BioAnalyzer. The Taqman Human Inflammation Array was used. A comparison of the amount of inflammatory gene expression in benchmark housekeeping gene 18S was made via quantitative PCR. The entire process was done in duplicate.
Quantitative Pcr Confirms Gene Array Expression Data of Select Genes
Prior to analysis all purified RNA was checked for quality on an Agilent BioAnalyzer. Samples meeting the quality criteria, an RNA Integrity Number (RIN) score of 8 or above were chosen for analysis. RNA samples were first tested for residual genomic DNA contamination by performing QPCR using the TWIST gene assay (which is able to detect genomic DNA) and 2ng of RNA as template. No amplification signal was seen (data not shown). RNA samples were then reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) following the manufacturer’s instructions. Taqman assays were set up in triplicate using TaqMan Gene Expression Master Mix (Applied Biosystems) and cDNA generated from 2ng of RNA for each sample following the recommended protocol. Reactions were then run on an Applied Biosystems 7900ht Sequence Detection System using the standard cycling conditions (50C for 2 min; 95C for 10 min; 40 cycles of 95C for 15 sec, 60C for 1 min). Fold change calculations were made using the delta-delta-Ct method. TaqMan Low Density Array (TLDA) Human Inflammation Panel assays (Applied Biosystems cat# 4378722) were run according to manufacturer’s instruction using 80ng of cDNA per card region. Reagents and cycling conditions were as described above. Data was analyzed using the Applied Biosystems RQ Manager software (v1.2.1).
Results
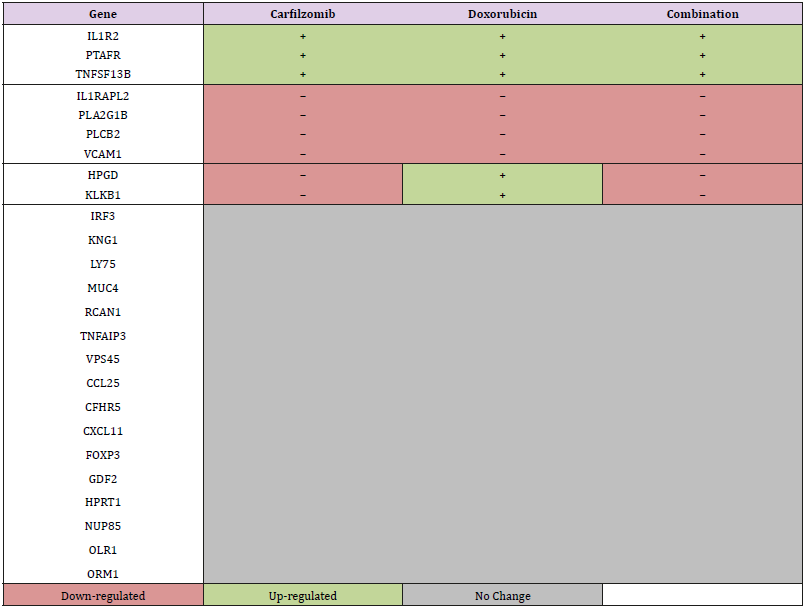
93 Inflammatory Gene Microarray design. The actions of inflammatory cytokines, growth factors, chemokines, transcription factors, prostaglandins, and leukotrienes are well recognized in both malignant and non-malignant states. Tables 1-2 display the selected genes for this study and also include control cDNAs of the housekeeping gene such as 18S for signal normalization and quantitation. Genes that were up-regulated by more than 1.5-fold or down-regulated by at least 1.5-fold were considered significant and a comparison of fold-change in expression of these controls, MCF-10A versus MDA-MB-231, was made (Tables 1A and 1B). In addition, a more comparative analysis and categorization of gene expression were made based on similarities and differences in individual expression profiles (Table 2).
In the MCF-10A cell line, PTAFR and IL1R2 expressed increased gene expression in duplicate in two different controls, while IL1RAPL2 showed down-regulation. The changes in expression observed were approximately 10-fold or greater. In the MDAMB- 231 cell line, TNFSF13B and VCAM1 were consistent in demonstrating decreased gene expression in duplicate in two different controls. Additionally, significant decreased expression of pro-inflammatory gene KLKB1 was also observed. Genes with significant increase, decrease, or no change in expression and whose data sets agreed with each other, were categorized according to the direction of fold-change (Table 2).
Discussion
In this experiment, we found many inflammatory genes to be up-regulated in the MDA-MB-231 TNBC cell line with distinct regulatory changes upon addition of doxorubicin and carfilzomib. Some of the resulting expression changes effectively reduced the inflammatory state, while other genes potentiated inflammation. It is well known that the production of these genes is important in promoting the activation of cytokines, angiogenesis, and metabolism of prostaglandins in breast tissue. Understanding the effects of carfilzomib and doxorubicin on inflammatory gene expression levels can help better elaborate the mechanisms behind these drugs and provide a comparative analysis of normal breast versus a TNBC inflicted state.
Carfilzomib Induces Down-Regulation of Pro- Inflammatory Gene Klkb1
Amongst the genes studied and targeted in our experiment, we observed the unexpected change in regulation of two novel genes previously unmentioned in the literature. The enzyme encoded by KLKB1 is known to be involved in a pro-inflammatory cascade, ultimately resulting in the release of Bradykinin Matsumoto et al. [9]. Bradykinin promotes inflammation by increasing blood vessel permeability and thus serves as a potential trigger for enhanced metastasis. HPGD, on the other hand, encodes an anti-inflammatory enzyme that metabolizes prostaglandin Lee et al. [10]. In our study, we only observed KLKB1 down regulation when MDA-MB-231 cells were given either standalone carfilzomib or combinational with doxorubicin. Doxorubicin alone resulted in an up regulation of KLKB1, likely leading to an increase in inflammatory effects. Contrastingly, HPGD was only up regulated with doxorubicin. The specific fold changes of HPGD and KLKB1 show that carfilzomibinduced KLKB1 down regulation was greater than the doxorubicininduced HPGD up regulation, again illustrating a potential for reduced inflammatory effects. However, further studies isolating and silencing KLKB1 and HPGD are needed to better elucidate their mode of action in TNBC.
Carfilzomib and Doxorubicin Induce Down-Regulation of Pro-Inflammatory Gene Vcam1
There were two specific genes that consistently showed increased expression in MDA-MB-231 cells; TNFSF13B and VCAM1. The former is a cytokine expressed in B cells and is also a potent activator Gene 2015b [11]. It has been found to be constantly expressed in adipocytes of normal breast cells and promotes the proliferation and development of normal as well as malignant breast tissue Pelekanou et al. [12]. The latter is an NF-kB related gene that is responsible for encoding a cell surface protein that mediates leukocyte-endothelial cell adhesion. Its up regulation is tumor-promoting, particularly in inflammatory breast cancer Lerebours et al. [13]. In addition, VCAM1 also plays a significant role in tumor cell metastasis through survival signals provided by metastasis-associated macrophages Lewis et al. [14]; O’Hanlon et al. [15]; Qian and Pollard [16]. Our investigation revealed that treatment of the MDA-MB-231 cell line with the second-generation proteasome inhibitor, carfilzomib, resulted in a decrease in gene expression for VCAM1, and that combining it with the first line TNBC chemotherapeutic agent, doxorubicin, seemed to decrease the amount of gene expression even further. On the other hand, gene expression of TNFSF13B was up regulated with the individual and combination of treatments. This suggests that a potential for an even greater reduction in inflammatory effects exists if TNFSF13B can be suppressed along with VCAM1. Since DOX/CARF interfere with NF-kB activation, a proposed mechanism of action could be the downstream deregulation of the CAM family in TNBC tumor cells, leading to defective angiogenesis and diminished metastasis Banks et al. [17].
Carfilzomib and Doxorubicin Induce Il1rapl2 Gene Suppression
In the MCF-10A normal breast epithelial cell line, we found consistently increased gene expression with PTAFR and IL1RAPL2. PTAFR serves as a binding site for platelet activating factor, which is a phospholipid that plays a significant role in inflammation, and its production in MDA-MB-231 cells has been correlated to angiogenesis, increased cell motility, and proliferation Bussolati et al. [18]; Gene 2015a [19]. IL1RAPL2 is a receptor for an interleukin 1 accessory protein that when activated, facilitates signal transduction pathways. IL-1 is a cytokine with various functions in both physiological and pathological states and is known to be up regulated in many tumor types including breast tumors Lewis et al. [14]. Treatment of the MDA-MB-231 cell line with carflizomib, doxorubicin, and a combination of both increased PTAFR gene expression, whereas the same treatments decreased IL1RAPL2 gene expression. The up-regulated expression of PTAFR presents a future potential target for a further enhanced inflammatory suppression. Regarding IL1RAPL2, the encoded receptor proteins in unaffected breast cells within a TNBC paradigm are targeted. Carflizomib, through proteasome inhibition, may up-regulate a binding moiety that binds preferentially to IL1RAP and diminishes its activity. This in turn would inhibit IL-1 dependent activation of NF-kB Dinesh Vyas MD [20], leading to diminished growth and inflammation. This, however, is only a proposed model of IL1RAP and drug interaction. As with VCAM1, the combined treatment resulted in a greater decrease in gene expression when compared to individual treatments. This suggests that down regulating expression of IL1RAPL2 with carfilzomib alongside doxorubicin could help further curb the pro-inflammatory processes.
Our investigation shows there are specific genes that have greater expression in the inflammatory response of MDA-MB-231 cells. This is supported and confirmed by multiple studies conducting gene profiles on inflammatory breast cancer subtypes Bertucci et al. [21]; Sorlie et al. [22]; Van Laere et al. [23]. Combined with our analysis of fold-changes after treatment with carfilzomib and doxorubicin, it is notable that inflammation levels can be decreased even further upon addition of the second-generation proteasome inhibitor. More anti-inflammatory results were seen with the down regulation of KLKB1, VCAM1, and IL1RAPL2, all of which are known to be up involved in TNBC (Vyas D) [24,25]. Similarly, targeting and minimizing the up-regulation of HPGD, TNFSF13B and PTAFR can potentially further reduce inflammation-induced toxicity and metastasis. The increased downregulation of genes seen with carfilzomib highlights its potential to be used either as a standalone or combinational drug alongside with doxorubicin. Further studies such as gene silencing or knockout are required to fully understand the role and mechanism of carfilzomib and doxorubicin on the TNBC inflammatory disease profile. However, it is evident that a supplementary or alternative strategy is necessary to ameliorate the negative effects of TNBC prognosis. Our microarray study serves as a preliminary insight towards an enhanced regimen for tumor-suppression in TNBC [26,27].
References
- Kanaya N, Adams L, Takasaki A, Chen S (2014) Whole blueberry powder inhibits metastasis of triple negative breast cancer in a xenograft mouse model through modulation of inflammatory cytokines. Nutr Cancer 66(2): 242-248.
- Mantovani A, Allavena P, Sica A, Balkwill F (2008) Cancer-related inflammation. Nature 454: 436-444.
- Lopez-Hisijos N C, LS, Shah P, Vyas A, Vyas D (2015) Manuscript for: A second-generation proteasome inhibitor is more potent than doxorubicin modulating IL-6, pSTAT-3 and NF-kB activity in triple negative breast cancer. Oncotarget.
- Stagg J, Allard B (2013) Immunotherapeutic approaches in triple-negative breast cancer: Latest research and clinical prospects. Ther adv med oncol 5: 169-181.
- Bandyopadhyay A, Wang L, Agyin J, Tang Y, Lin S, et al. (2010) Doxorubicin in combination with a small TGFbeta inhibitor: a potential novel therapy for metastatic breast cancer in mouse models. PloS one 5: e10365.
- Vyas D, Lopez-Hisijos N, Gandhi S, El-Dakdouki M, Basson MD, et al. (2015) Doxorubicin-Hyaluronan Conjugated Super-Paramagnetic Iron Oxide Nanoparticles (DOX-HA-SPION) Enhanced Cytoplasmic Uptake of Doxorubicin and Modulated Apoptosis, IL-6 Release and NF-kappaB Activity in Human MDA-MB-231 Breast Cancer Cells. J Nanosci Nanotechnol 15(9): 6413-6422.
- Riz I, Hawley TS, Hawley RG (2015) KLF4-SQSTM1/p62-associated prosurvival autophagy contributes to carfilzomib resistance in multiple myeloma models. Oncotarget 6: 17814-14831.
- Vyas D, Gambhir S, Hollis M, Aekka A, Vyas A (2015) Nuclear factor kappa B role in inflammation associated gastrointestinal malignancies. World J gastroenterol 21: 3174-3183.
- Matsumoto H, Koo SL, Dent R, Tan PH, Iqbal J (2015) Role of inflammatory infiltrates in triple negative breast cancer. J clinical pathol 68: 506-510.
- Lee MY, Tse HF, Siu CW, Zhu SG, Man RY, et al. (2007) Genomic changes in regenerated porcine coronary arterial endothelial cells. Arterioscl thromb vascul biol 27: 2443-2449.
- Gene N (2015b) TNFSF13B tumor necrosis factor (ligand) superfamily, member 13b Homo sapiens (human).
- Pelekanou V, Kampa M, Kafousi M, Darivianaki K, Sanidas E, et al. (2008) Expression of TNF-superfamily members BAFF and APRIL in breast cancer: immunohistochemical study in 52 invasive ductal breast carcinomas. BMC cancer 8: 76.
- Lerebours F, Vacher S, Andrieu C, Espie M, Marty M, et al. (2008) NF-kappa B genes have a major role in inflammatory breast cancer. BMC cancer 8: 41.
- Lewis AM, Varghese S, Xu H, Alexander HR (2006) Interleukin-1 and cancer progression: The emerging role of interleukin-1 receptor antagonist as a novel therapeutic agent in cancer treatment. J trans med 4: 48.
- O'Hanlon DM, Fitzsimons H, Lynch J, Tormey S, Malone C, et al. (2002) Soluble adhesion molecules (E-selectin, ICAM-1 and VCAM-1) in breast carcinoma. Eur J cancer 38: 2252-2257.
- Qian BZ, Pollard JW (2012) New tricks for metastasis-associated macrophages. Breast cancer res 14: 316.
- Banks RE, Gearing AJ, Hemingway IK, Norfolk DR, Perren TJ, et al. (1993) Circulating intercellular adhesion molecule-1 (ICAM-1), E-selectin and vascular cell adhesion molecule-1 (VCAM-1) in human malignancies. Br J cancer 68: 122-124.
- Bussolati B, Biancone L, Cassoni P, Russo S, Rola-Pleszczynski M, et al. (2000) PAF produced by human breast cancer cells promotes migration and proliferation of tumor cells and neo-angiogenesis. The Am J pathol 157: 1713-1725.
- Gene N (2015) PTAFR platelet-activating factor receptor Homo sapiens (human).
- Vyas D LG, Vyas AK (2014) Chemotherapy-enhanced inflammation may lead to the failure of therapy and metastasis. Onco Targets Ther 7:1015-1023
- Bertucci F, Finetti P, Rougemont J, Jauffret EC, Cervera N, et al. (2005) Gene expression profiling identifies molecular subtypes of inflammatory breast cancer. Cancer resea 65: 2170-2178.
- Sorlie T, Wang Y, Xiao C, Johnsen H, Naume B, et al. (2006) Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: Gene expression analyses across three different platforms. BMC genomics 7: 127.
- Van Laere SJ, Van den Eynden GG, Van der Auwera L, Vandenberghe M, Van Dam P, et al. (2006) Identification of cell-of-origin breast tumor subtypes in inflammatory breast cancer by gene expression profiling. Bre can res treat 95: 243-255.
- Cao Y, Karin M (2003) NF-kappaB in mammary gland development and breast cancer. J mammary gland biol neoplasia 8: 215-223.
- de Oliveira SI, Andrade LN, Onuchic AC, Nonogaki S, Fernandes PD, et al. (2010) Platelet-Activating Factor Receptor (PAF-R)-dependent pathways control tumour growth and tumour response to chemotherapy BMC cancer 10: 200.
- Fisher B, Anderson S, Tan-Chiu E, Wolmark N, Wickerham DL, et al. (2001) Tamoxifen and chemotherapy for axillary node-negative, estrogen receptor-negative breast cancer: findings from National Surgical Adjuvant Breast and Bowel Project B23. J clin oncol 19: 931-942.
- Normanno N, De Luca A, Carotenuto P, Lamura L, Oliva I, et al. (2009) Prognostic applications of gene expression signatures in breast cancer. Oncology 1: 2-8.

 Research Article
Research Article