Abstract
In this study 64 yeast isolates were isolated from natural sources, out of them 6 red yeasts were selected and named as AUN-F1, AUN-F4, AUN-F5, AUN-F7, AUN-F38 and AUN-F55. The selected isolates were screened for their antagonistic property against pathogenic bacteria that caused postharvest diseases. The isolate AUN-F55 recorded the highest inhibition zone against Erwinia carotovora it was 12.67mm and isolate AUN-F5 was the lowest with inhibition zone 8.67mm. The isolate AUN-F7 recorded the lowest inhibition zone against Erwinia chrysanthemi it was 10.33mm. RAPD-PCR fingerprinting was carried out for the six isolates, and the DNA patterns revealed that there is no correlation between the RAPD profile and geographic origin sites where these isolates were collected from.
Keywords: Rhodotorula Spp; Antagonistic; Erwinia Carotovora; Erwinia Chrysanthem; RAPD-PCR fingerprinting
Introduction
Rhodotorula is a genus of yeasts, color classified in the phylum Basidiomycota, family Sporidiobolaceae, subfamily of Rhodotorulalodeae and class Microbotryomycetes. These are found from widely varying environmental sources ranging from extreme climates of deep-sea vents to arctic cold deserts [1-3] from the oil, air, or as organisms associated with plants. All of Rhodotorula yeasts produce carotenoids, and their colonies develop a characteristic salmon pink color [4, 5]. Rhodotorula species also play a role in the market of biocontrol agents, also known as antagonistic yeast. Significant losses in harvested fruit occur from decay by filamentous fungi such as Botrytis cinerea and Penicillium expansum [6]. In order to replace or augment chemical fungicides, biological control agents have been studied heavily the past two decades, with some yeast-based biocontrol products commercially available for specific commodities [7]. The ability to control different rots, on different fruits/vegetables, is still an important goal when developing postharvest biocontrol products. Therefore, antagonistic yeasts continue to be an active area of research, and several Rhodotorula species have shown promise in this field. Therefore, this study aims to isolate and purify of yeast strains from natural sources, evaluate and screen of red yeast isolates for control of postharvest pathogens bacteria. Finally, discrimination at the molecular level within selected red yeast isolates using random amplified polymorphic DNA (RAPD)-PCR fingerprinting was determined.
Materials and Methods
Sampling, Yeast Isolation and Purification
Fruit, vegetable, juices and soil samples were collected from different farms and local markets in Assiut, Egypt. Isolation was performed on autoclaved yeast extract peptone dextrose (YEPD) agar according to [8] One gram of each sample was placed in a conical flask containing 50mL sterile distilled water and shaken for 30 min. Then, 0.1mL aliquots of the suspension were spread on agar plates and incubated at 28 °C for 48-72h. After incubation, individual colonies were selected randomly according to their different color and morphological characteristics and were purified by single colony isolation after triple restreaking on the same medium. Finally, each pure culture was preserved in slants of the medium at 4 °C for further use [9].
Pathogenic Bacteria
Pathogenic bacteria were brought from the Department of Plant Pathology, Faculty of Agriculture, Assiut University, Egypt.
Antimicrobial Activity Assay
YPDA medium solidified with 1.0% agar was used for the agar diffusion assay. The medium pH was adjusted to 5.0 or 6.0. After autoclaving at 121 1C for15 min, the medium was cooled to 40 1C in a water bath. One of each bacteria was cultivated into 15ml YPDA medium and was incubated at 28 °C for 24h, then, they were swept separately onto YPDA plates using sterile swabs. From the previously prepared yeast agar plates, few yeast colonies were carefully picked up using platinum loop and spotted at the pathogen-inoculated plates. The plates were incubated at 27 C for 48-72 h and the width of the inhibition zones was measured. Sterile water spotted in the plates with the pathogen was used as control.
RAPD-PCR Fingerprinting
Isolation of Genomic DNA from Yeast
The extraction of total yeast genomic DNA was performed according to the procedures described by Hesham [10].
Polymerase Chain Reaction (PCR) Amplification for RAPD
To study the genetic differences and relationships among six yeast isolates, five RAPD-PCR primers OPA-2, OPA-3, OPA-9, OPA-11 and OPA-15 were used [11]. PCR was performed in a final volume of 25μl containing GoTaq (Promega, Madison, WI, USA) green master mix, 1μl DNA sample and 2.0μl primer. The amplification reactions were carried out in a thermocycler under the following conditions for RAPD-PCR: initial denaturation at 94 ºC for 5 min, followed by 45 cycles of 92 ºC for 1 min, 36 ºC for 1 min, 72 ºC for 2 min, and a final extension at 72 ºC for 10 min and subsequently cooled to 4 ºC. Five μl of the amplified products were then analyzed using 1.5% 0.5×TBE agarose gel electrophoresis. The gel was stained with ethidium bromide 0.01%, visualized under UV light and photographed. The molecular sizes of DNA fragments were obtained with comparison with a 100-bp molecular marker.
RAPD Analysis
All the fragments generated by RAPD-PCR using the primers were considered and analyzed separately for determining the band types. The size of each band was determined using Kodak Digital Science 1D (KSD1D 2.0, Rochester, USA) software and the presences (1) or absences (0) of a band were recorded to generate a binary table. The data tables were export into the NTSYS-pc software [12] for analysis. The normalized RAPD patterns were further analyzed using Gel compare II version 2.5 (Applied Maths, St Martens-Latem, Belgium).
The levels of similarity in the RAPD was calculated using Pearson’s correlation coefficient. Cluster analysis was performed using the unweighted pair group method with arithmetic mean (UPGMA) method. The relationships between the pattern profiles are displayed as dendrograms and expressed as percentage similarity. The numerical discriminatory index (D) which is a measure of the discriminatory ability of the typing methods was calculated according to [13].
Statistical Analysis
Each treatment was carried out 3 replications. The experiment designed with randomized complete design. All the data were subjected to Analysis of Variance (ANOVA) using Cost at package version 6.311. The means were compared according to Duncan’s multiple range tests at P ≤0.05 [14].
Results
Yeast Isolation and Characterization
In this study 64 yeast strains were isolated from different natural sources in the Assiut region, Egypt, and purified by single colony isolation after triple re-streaking on YEPD agar plates, out of them 6 red yeast. Colony surface (smooth), margin (undulating), elevation (convex), and color (red) of the yeast isolates were recorded.
Antimicrobial Activity Test
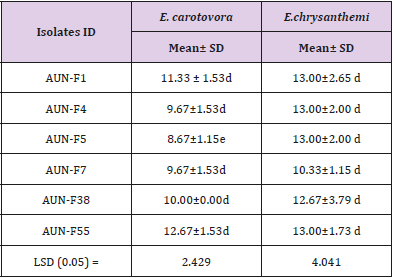
All yeast isolates were screened by agar diffusion assay for their antagonistic property against E-rwinia carotovora and Erwinia chrysanthemi. 36 out of the 64 isolates (56. 25 %) Inhibited E. chrysanthemi growth and 25 out of the 64 isolates (39.06%) Inhibited the growth of E. carotovora (Figures 1-2) all genera identified induced a clear inhibition zone, which indicated that these yeast isolates exhibited antagonistic activity. Out of them 13 have higher IZ values. Six of 13 yeast isolates were Rhodotorula ssp (Table 1). The others failed to induce antisporulant activity on bacteria.
Table 1: The inhibition zone of Rhodotorula ssp. IZ indicates the inhibition zones between the yeasts and the bacteria. The results are presented as the mean ± standard deviation.
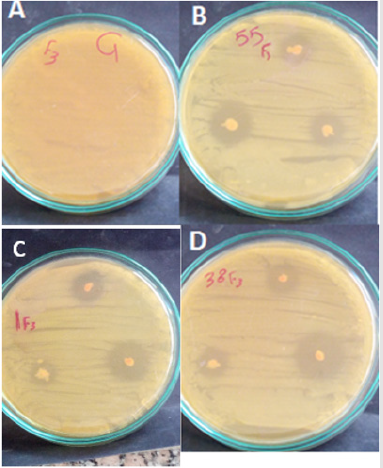
Figure 1: In vitro test of antagonism of red yeast isolate towards E. chrysanthemi using the spot method on YPDA plates. Pictures taken 24 hours after incubation of yeast with the pathogen. (A)Control E.chrysanthemi, (B,C and D) Rhodotorula ssp.
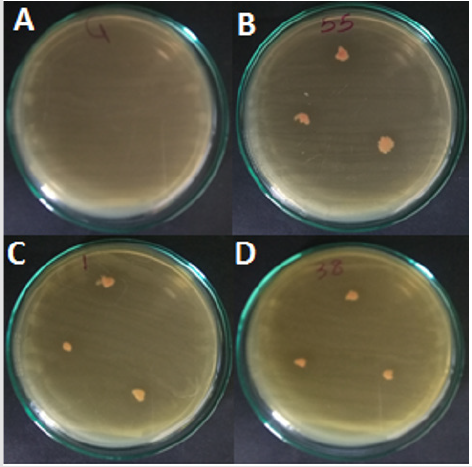
Figure 2: In vitro test of antagonism of yeast isolate towards E. chrysanthemi the spot method on YPDA plates. Pictures taken 24 hours after incubation of yeast with the pathogen. (A) Control E carotovora, (B, C and D) Rhodotorula ssp.
Genetic Differentiation by RAPD-PCR Fingerprinting Technique
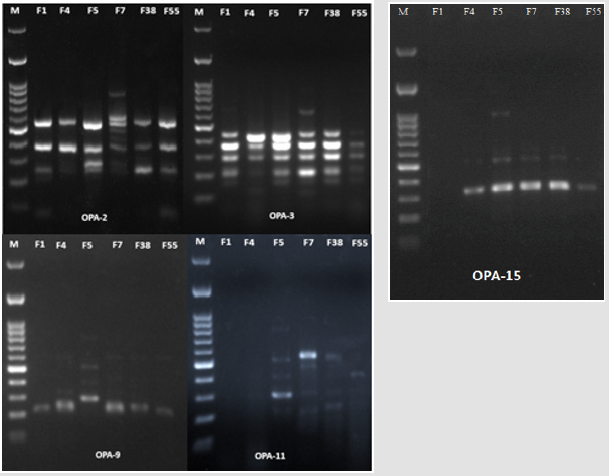
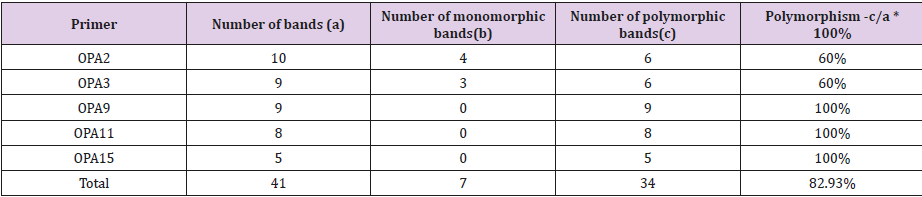
The six antagonistic red yeast strains were selected for analysis by RAPD-PCR fingerprinting to examine differentiation at the molecular level using the five random ten-mer primers. The results elucidated that the number of amplified fragments for each primer varied between 5 and 10 fragments, as in Table 2, and their sizes ranged from 40 to 1400bp based on Figure 3, Table 2 showed also that the total number of bands was 3, whereas the polymorphism percentage was (82.93%).
Figure 3: Agarose gel electrophoresis of RAPD products by OPA-2, OPA-3, OPA-9, OPA-11 and OPA-15 primers..
Genetic Similarity Matrix and Cluster Analysis
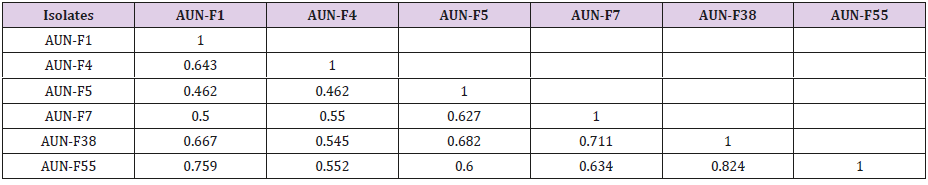
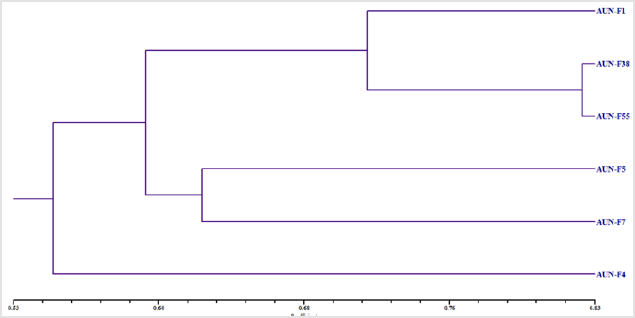
Data of the presence / absence of DNA fragments (markers) phenotypically analyzed using MVSP program of [15], and pairwise comparisons between the tested isolates of yeast were used to calculate the genetic similarity. Then, based on the calculated genetic similarity presented in Table 3. The results in Table 3, revealed that the lowest genetic similarity was noticed between strains AUN-F1 and AUN-F4 with AUN-F5 (46.2%), while the highest rate was noticed between strains AUN-F38 and AUN-F55 (82.4%). Consequently, all six strains of red yeasts were regrouped in three main clusters (Figure 4). Based on the dendrogram in Figure 4, three strains, AUN-F1, AUN-F38 and AUN-F55 were included in one cluster, while isolates AUN-F5 and AUN-F7 were in the other cluster. Contrastingly, AUN-F4 was placed in a separate cluster.
Table 3: Genetic similarity values calculated from the DNA fragments amplified from six red yeast isolates using five RAPD primers.
Figure 4: Dendrogram demonstrating the relationship among the sex isolates of yeast AUN-F1, AUN-F4, AUN-F5, AUN-F7, AUN-F38 and AUN-F55 based on data recorded from polymorphism of RAPD markers.
.Discussion
In this study, we investigated the inhibition effects of red yeast (Rhodotorula spp.) antagonists against E. carotovora and E. chrysanthemi, that causal agent of soft mold disease of potatoes, onions, carrots and other vegetables. An antagonistic Rhodotorula strain has been reported as an effective biocontrol agent against postharvest decay of apples, pears, sweet cherries, and oranges. Rhodotorula glutinis is an effective biocontrol agent against postharvest fungal spoilage of apples [16] pears [17] oranges [18] sweet cherries [19] as well as strawberries [20]. Biocontrol efficacy was enhanced with the addition of salicylic acid. Rhodotorula mucilaginosa is a strain of antagonistic yeast which was isolated from the soil sample of an unsprayed orchard by our research team. Our research team found that R. mucilaginosa showed biocontrol efficacy against blue mold and gray mold of apples caused by P. expansum and B. cinerea respectively [21]. The phylloplane yeast Rhodotorula mucilaginosa has been reported to control B. cinerea on geranium seedlings in combination with fungicides [22]. Rhodotorula minuta is reported as an antagonistic microorganism of the pathogen C. gloeosporioides. The proposed antagonistic mechanism of R. minuta is by competition for space and limited resources as no inhibition zone was observed near to the antagonistic colonies [23]. Biocontrol can also be applied by means of ‘killer’ activity, since R. glutinis and R. mucilaginosa have these effects on other microorganisms. Species representative of Rhodotorula are known to have ‘killer’ activity against most ascomycetous and basidiomycetous species. In addition, R. glutinis produces antibacterial compounds inhibitory to both Pseudomonas fluorescens and Staphylococcus aureus. Rhodotorula colostri produces and excretes an extracellular toxin, mycocin, which is lethal to other sensitive yeast strains [24].
For genetic differentiation, RAPD analysis was applied with the six red yeast strains (AUN-F1, AUN-F4, AUN-F5, AUN-F7, AUN-F38 and AUN-F55). RAPD analysis compares the whole chromosomal DNA displaying bands of genotypically specific values and is fast and trustworthy [25]. This technique uses short (5-10 mer) oligonucleotide primers with arbitrary sequences at low annealing temperatures that hybridize at loci distributed randomly throughout the genome, allowing the amplification of polymorphic DNA fragments [26]. RAPD results displayed various RAPD patterns, which indicated that the 6 yeasts: AUN-F1, AUN-F4, AUN-F5, AUN-F7, AUN-F38and AUN-F55 were various from each other could be distinctly separated by their RAPD patterns. According to [18], OPA-2, OPA-3, and OPA-9 were the best primers to discriminate yeast strains. The RAPD-PCR technique has been to distinguish between strains or subspecies of microorganisms [27]. Although the isolates AUN-F4 and AUN-F5 were obtained from the same location (Sahel slim), the dendrogram and genetic similarity regrouped these isolates in different clusters that were distinguished from each other. By contrast, despitestrains AUN-F5 and AUN-F7 being isolated from different locations (Sahel slim and Assiut city respectively), the results regrouped these yeasts in the same cluster. Therefore, there was no correlation between the RAPD patterns and the geographical origin of these strains which was in agreement with our previous work in bacterial isolates [28]. Our results using RAPD markers are similar to those previously reported [29,30]. They showed that the RAPD-PCR technique is useful for yeast strain identification, discrimination and estimation of the genetic diversity [31].References
- Vartivarian SE, Anaissie EJ, Bodey GP (1993) Emerging fungal pathogens in immunocompromised patients: classification, diagnosis, and management. Clinical infectious diseases 17(2): S487-S491.
- Margesin R, Fonteyne PA, Schinner F, Sampaio JP (2007) Rhodotorula psychrophila sp. nov., Rhodotorula psychrophenolica sp. nov. and Rhodotorula glacialis sp. nov., novel psychrophilic basidiomycetous yeast species isolated from alpine environments. International journal of systematic and evolutionary microbiology 57(9): 2179-2184.
- Zalar P, Gunde Cimerman N (2014) Cold-adapted yeasts in Arctic habitats. In Cold-adapted Yeasts Springer: p. 49-74.
- Wirth F, Goldani LZ (2012) Epidemiology of Rhodotorula: an emerging pathogen. Interdisciplinary perspectives on infectious diseases.
- Chattopadhyay P, Chatterjee S, Sen SK (2008) Biotechnological potential of natural food grade biocolorants. African Journal of Biotechnology 7(17).
- Castoria R, De Curtis F, Lima G, De Cicco V (1997) β-1, 3-glucanase activity of two saprophytic yeasts and possible mode of action as biocontrol agents against postharvest diseases. Postharvest Biology and technology 12(3): 293-300.
- Liu J, Sui Y, Wisniewski M, Droby S, Liu Y (2013) Utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. International journal of food microbiology 167(2): 153-160.
- Kurtzman CP, Fell JW, Boekhout T. Definition, classification and nomenclature of the yeasts, Elsevier.
- Hesham AEL, Alrumman SA, Al Dayel MA, Salah HA (2017) Screening and genetic identification of acidic and neutral protease-producing yeasts strains by 26S rRNA gene sequencing. Cytology and Genetics 51(3): 221-229.
- Hesham A (2014) New safety and rapid method for extraction of genomic DNA from bacteria and yeast strains suitable for PCR amplifications. J. Pure Appl. Microbiol 8(1): 383-388.
- Martorell P, Fernandez Espinar MT, Querol A (2005) Molecular monitoring of spoilage yeasts during the production of candied fruit nougats to determine food contamination sources. International journal of food microbiology 101(3): 293-302.
- Rohlf FJ (2000) NTSYS-pc Numerical taxonomy and multivariate analysis system. Applied Biostatistics.
- Hunter PR, Gaston MA (1988) Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity JCM 26(11): 2465-2466.
- Duncan DB (1955) Multiple ranged multiple F-test. Biometrics 11: 1- 47.
- Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences 76(10): 5269-5273.
- Qin GZ, Tian SP, Liu HB, Xu Y (2003) Biocontrol efficacy of three antagonistic yeasts against Penicillium expansum in harvested apple fruits. Acta Botanica Sinica Chinese Edition 45(4): 417-421.
- Zhang H, Wang S, Huang X, Dong Y, Zheng X (2008) Integrated control of postharvest blue mold decay of pears with hot water treatment and Rhodotorula glutinis. Postharvest Biology and Technology 49(2): 308-313.
- Zheng XD, Zhang HY, Sun P (2005) Biological control of postharvest green mold decay of oranges by Rhodotorula glutinis. European Food Research and Technology 220(3-4): 353-357.
- Tian SP, Qin GZ, Xu Y, Wang YS (2004) Application of antagonistic yeasts under field conditions and their biocontrol ability against postharvest diseases of sweet cherry. Acta Botanica Sinica English Edition 46(11): 1324-1330.
- Zhang H, Wang L, Dong Y, Jiang S, Cao J, et al. (2007) Postharvest biological control of gray mold decay of strawberry with Rhodotorula glutinis. Biological Control 40(2): 287-292.
- Li RP, Zhang HY, Liu WM, Zheng XD (2011) Biocontrol of postharvest gray and blue mold decay of apples with Rhodotorula mucilaginosa and possible mechanisms of action. International Journal of Food Microbiology 146: 151-156.
- Buck JW (2004) Combinations of fungicides with phylloplane yeasts for improved control of Botrytis cinerea on geranium seedlings. Phytopathology 94(2): 196-202.
- Patino Vera M, Jimenez B, Balderas K, Ortiz M, Allende R, et al. (2005) Pilot‐scale production and liquid formulation of Rhodotorula minuta, a potential biocontrol agent of mango anthracnose. Journal of Applied Microbiology 99(3): 540-550.
- Albertyn J, Pohl CH, Viljoen BC (2014) Rhodotorula. pp. 291-295.
- Molnar E, Baude A, Richmond SA, Patel PB, Somogyi P, et al. (1993) Biochemical and immunocytochemical characterization of antipeptide antibodies to a cloned GluR1 glutamate receptor subunit: cellular and subcellular distribution in the rat forebrain. Neuroscience 53(2): 307-326.
- Quesada M P, Cenis JL (1995) Use of random amplified polymorphic DNA (RAPD-PCR) in the characterization of wine yeasts. American Journal of Enology and Viticulture 46(2): 204-208.
- Martorell M, Fernandez Espinar MT, Querol A (2005) Molecular Monitoring of Spoilage Yeasts During The Production of Candied Fruit Nougats to Determine Food Contamination Sources. Int J Food Microbiol 101: 293-302.
- Tofalo R, Chaves Lopez C, Di Fabio F, Schirone M, Felis GE, et al. (2009) Molecular identification and osmotolerant profile of wine yeasts that ferment a high sugar grape must. International journal of food microbiology 130(3): 179-187.
- Hesham AEL, Alrumman SA, Al Amari JA (2016) 16S rDNA Phylogenetic and RAPD–PCR Analyses of Petroleum Polycyclic Aromatic Hydrocarbons-Degrading Bacteria Enriched from Oil-Polluted Soils. Arabian Journal for Science and Engineering 41(6): 2095-2106.
- Meroth CB, Hammes WP, Hertel C (2003) Identification and population dynamics of yeasts in sourdough fermentation processes by PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol 69(12): 7453-7461.
- EL Fiky ZA, Hassan, GM, Emam AM (2012) Quality Parameters and RAPD‐PCR Differentiation of Commercial Baker's Yeast and Hybrid Strains. Journal of food science 77(6): M312-M317.

 Research Article
Research Article