Abstract
Purpose: To develop an accurate, automated multi-atlas segmentation algorithm for creating three-dimensional sigmoid sinus models from clinical computed tomography (CT) volumes for use in temporal bone mastoidectomy surgical simulation software.
Methods: Clinical CT and micro-CT scans of 38 cadaveric temporal bones were used to develop and validate the algorithm. A single-atlas and multi-atlas segmentation were compared for accuracy using three different label fusion methods: majority voting, STAPLE, and joint label fusion. The automated segmentation algorithm was evaluated by comparing to ground truth manual segmentations through a combination of visual inspection and Dice, Hausdorff distance, and average Hausdorff distance metrics.
Results: The best results were obtained for multi-atlas segmentation using joint label fusion for which a mean Dice value of 0.77 was found across all samples when compared to the manual segmentations. The mean Hausdorff distance was 10.39 mm, and the mean average Hausdorff distance was 0.30 mm, corresponding to less than two voxels. Visual inspection revealed accurate and high-resolution segmentations.
Conclusion: The presented multi-atlas method is effective and accurate at automatically producing high-resolution segmentations of the sigmoid sinus for the purpose of surgical simulation.
Keywords: Sigmoid sinus; Automatic segmentation; Atlas-based segmentation; Temporal bone anatomy
Introduction
Mastoidectomy is a complex surgical procedure that requires extensive knowledge of the anatomy of the temporal bone and is often required in cochlear implantation surgery. However, due to its complexity, mastoidectomy is a difficult procedure for trainees to master. Traditional training methods for surgical residents utilize cadavers, which can be expensive and difficult to access. To provide more consistent and accessible training for surgery involving the temporal bone, surgical simulators have been developed that provide haptic (touch) feedback and three-dimensional (3D) visualization [1-10]. Surgical simulation is becoming a widely accepted tool in Otolaryngology since it offers the ability to model difficult and varied cases and allows trainees to practice on patientspecific models.
Simulators such as CardinalSim [10] are able to import patient images and can be used for both training and in pre-operative planning. To maximize the variety and relevance of the anatomical cases available in the simulator, many clinical scans need to first be segmented (i.e., the anatomy needs to be delineated) for use. A major drawback to VR simulators is the need for manual image segmentation, which can take hours per scan and is infeasible in a clinical setting. Automated segmentation methods are preferred to be able to rapidly and automatically produce a variety of 3D digital models of the temporal bone. One challenge associated with the development of simulators in Otolaryngology is the complex anatomy of the temporal bone. For example, in mastoidectomy one of the vital anatomical structures is the sigmoid sinus. The sigmoid sinus is a venous sinus that travels down an S-shaped groove in the temporal bone. During the initial portion of the procedure the sigmoid sinus represents the posterior boundary of bone removal [11]. It is critical for surgeons and trainees to be able to identify the sigmoid sinus to avoid catastrophic vascular complications during surgery.
Currently, creating 3D models of the sigmoid sinus requires manual delineation of structural boundaries by an individual with expertise in both the anatomy and the software tools used to segment medical image volumes. In addition, the sigmoid sinus is a highly variable structure in both shape and relative position to other structures in the temporal bone, and previous work has focused on evaluating its variability through statistical shape analysis [12]. Due to this variability, it is very time-consuming for an expert to manually perform segmentation (often taking up to 45 minutes). Therefore, an automated algorithm is required to produce sigmoid sinus segmentations accurately and quickly with minimal expert intervention. However, due to the vast anatomic variability and low contrast at the medial wall, purely intensity-based methods such as thresholding are inconsistent, making development of an automated segmentation method of the sigmoid sinus uniquely challenging.
Several approaches to automated segmentation of anatomical structures have been described in the literature. One of the simplest approaches is thresholding. Thresholding is a fast and effective method for delineating structures that have high contrast from the surrounding objects, but as noted above this is generally ineffective for many anatomical structures such as the sigmoid sinus which has similar intensity values to its surrounding areas on its medial side. Atlas-based methods are far more promising for capturing variability in difficult to delineate anatomical structures due to their ability to capture the anatomical information and their relative robustness to poor contrast. One effective application of atlasbased segmentation on various structures of the temporal bone that excludes the sigmoid sinus has been presented by Powell et al. [13]. The present work describes the development and evaluation of a multi-atlas-based segmentation algorithm that compares a variety of label fusion methods on clinical CT scans of cadaveric temporal bones with the goal of accurately segmenting the sigmoid sinus. The multi-atlas method presented herein has been applied previously to segment medical images in a variety of fields [14- 16] and has been adapted in this work to allow for the usage of highly detailed micro-CT (μCT) atlases to segment low resolution and poorly delineated clinical CT scans. This method was able to capture the high variability of the anatomy of the sigmoid sinus.
Materials and Methods
Data
Thirty-eight anonymized adult cadaveric temporal bones with normal anatomy were used. Samples had not been operated on in any previous surgeries and were scanned using a General Electric (GE) Healthcare explore Locus μCT scanner at a resolution of 154μm x 154μm x 154μm and a voltage of 80kV. Clinical-CT scans of the same thirty-eight samples were also collected using a resolution of 234μm x 234μm x 625μm and a voltage of 120kV on a Discovery CT750 HD Clinical Scanner with GE’s Gemstone CT detector. All cadaveric specimens were obtained with permission from the body bequeathal program at Western University, London, Ontario, Canada in accordance with the Anatomy Act of Ontario and Western’s Committee for Cadaveric Use in Research (approval number: #19062014).
Ground Truth Segmentations and Atlas Creation
The sigmoid sinus was manually segmented from the μCT images by an expert anatomist (KV) using a combination of semi-automated and manual tools in 3D Slicer, an open-source software for medical image processing and visualization [17]. Consensus interpretation of the segmentations was achieved by an experienced surgeon (SKA), the anatomist (KV), and the lead author (DA). Details of the segmentations and anatomic analysis have been previously described in [12]. Twelve segmentations were used to define atlases to drive the segmentation algorithm and the remainder formed the ground truth and were used to evaluate the segmentation algorithm. Two sets of atlases were defined from the 12 segmentations: 6 atlases for left temporal bones and 6 for right temporal bones. This number of atlases managed to capture the variability of the sigmoid sinus while producing results comparable to using 36 atlases. The use of μCT images resulted in higher resolution and detail for both the ground truth and the algorithm-generated segmentations. As the algorithm is applied to clinical-CT volumes, the μCT segmentations were registered using a combination of rigid and affine techniques to their corresponding clinical-CT volumes and then reviewed by the research team to be used as a high-resolution ground truth.
Segmentation Algorithm
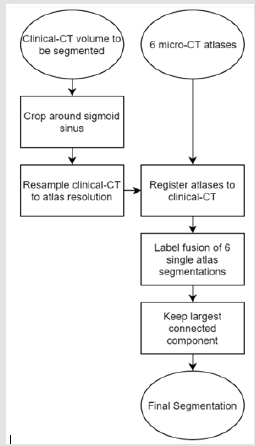
Figure 1 depicts the operation of the segmentation algorithm. All steps are completely automatic with the exception of basic rough cropping of the image. The major steps of the algorithm are described next in further detail. The algorithm was implemented as one single script written in Bash shell script.
Figure 1: Block diagram displaying the multi-atlas segmentation algorithm. User specifies either left or right ear and appropriate atlases are chosen by algorithm.
Cropping and Resampling: The clinical-CT volumes were cropped around the area of the sigmoid sinus and resampled using linear interpolation from 0.234 mm x 0.234 mm x 0.625 mm to 0.154 mm x 0.154 mm x 0.154 mm isotropic to be approximately the same resolution as the μCT volumes. This was done to reduce the loss of detail from the μCT atlas segmentations during registration and resampling to the resolution of the target clinical-CT volumes.
Rigid and Affine Registration: Prior to non-rigid registration, a rigid step and affine step were used to approximately align each atlas to the target clinical-CT volume. This two-step approach was taken to improve accuracy. By applying the rigid registration first, the time required for the affine transformation is reduced, approximate before non-rigid registration. This two-step registration approach was performed using the NiftyReg implementation of a symmetric (source to target and target to source, simultaneously) blockmatching registration, applied in three pyramidal levels from coarse to fine, doubling the resolution on each step up to the original 0.154 mm isotropic voxel size [18].
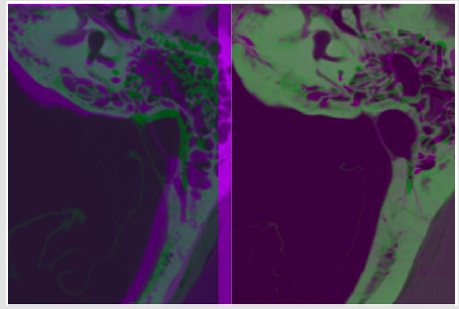
Non-Rigid Registration: Non-rigid registration for the μCT atlases was also accomplished using the NiftyReg implementation of a B-spline pyramidal approach in three progressively finer control point grids using 12mm x 12mm x 12mm, 6mm x 6mm x 6mm, and 3mm x 3mm x 3mm grid spacings, respectively [19]. The loss function used by NiftyReg for the non-rigid registration was a combination of normalized mutual information (NMI) and bendingenergy (BE), which was optimized using a conjugate gradient scheme. An example of the process of volume registration from rigid to the final non-rigid B-spline registration is given in Figure 2.
Figure 2: Axial images of the temporal bone showing target clinical CT (magenta) and the μCT atlas (green). The unregistered images are shown overlaid without registration (left) and the result of a complete registration (right). Observe the shift of the position and shape of the sigmoid sinus.
Label Fusion: Individually, the accuracy when comparing Dice and Hausdorff distance values of the results of the single-atlas segmentations varied, but consistency and overall accuracy were greatly improved by combining the information from each of the six single-atlas segmentations for a given clinical-CT volume using label fusion methods. Three established and widely available label fusion methods were applied to the completed registrations: majority voting, STAPLE [20], and joint label fusion [14]. The registrations were then compared to determine the differences between the methods as they related to the sigmoid sinus.
A. Majority Voting: The first label fusion method applied was majority voting. Majority voting counts the value (either 0 or 1) of each individual binary segmentation at each voxel in the image volume and takes the majority decision as the result. This method is fast and simple to apply and improves the consistency of the segmentation quality when compared to single-atlas segmentations. A disadvantage of this method is that inaccurate and outlying segmentations are given the same weight as the more accurate segmentations.
B. STAPLE: STAPLE is an expectation maximization algorithm for evaluating the performance of multiple separate segmentations and produces a final probabilistic segmentation. As opposed to majority voting, STAPLE aims to use the data from all the individual segmentations to determine performance levels of each individual segmentation and then uses that information to find a final segmentation deemed closest to the true segmentation by the algorithm [20]. On average, STAPLE produces much better results compared to majority voting; however, it takes more time especially when evaluating multiple atlases.
C. Joint Label Fusion: The third and final label fusion method used was the Advanced Normalization Tools (ANTs) implementation of joint label fusion. Joint label fusion adopts a similar statistical approach to STAPLE, but also uses information from each registration image volume result along with the generated label map. Joint label fusion applies the probability that multiple atlases would make the same error at a particular voxel [14].
D. Largest Connected Component: After the multi-atlas procedure was completed with label fusion, island removal was performed to remove noise and disconnected voxels from the segmentation usually caused by the label fusion methods. Largest connected component island removal is quick to apply and has been applied in previous segmentation applications [13]. Since the sigmoid sinus is one connected blood vessel, the approach used for island removal was to discard all but the largest connected component of the segmentation. This resulted in a clean single label with no noise that was ready for use in a surgical simulator. Evaluation and Metrics
The automated segmentations generated by the multi-atlasbased method were evaluated by comparing to the ground truth, manual segmentations completed by the anatomist using a variety of metrics. The segmentation algorithm was applied to clinical-CT volumes, but the assessment was done with comparison to ground truth μCT which were registered to their corresponding clinical-CT since label maps are higher resolution in μCT as well as boundaries being more visible in μCT. The first metric used was the Dice coefficient, which determines the overlap between the automated and manual segmentations. The second metric used was the Hausdorff distance, which measures the maximum distance from one segmentation to another. The Hausdorff distance is extremely sensitive to noise, such that algorithms which segment a smaller or larger portion of the structure than the ground truth will return larger values while segmenting the correct areas.
Island removal completed in post-processing is reasonably effective at negating this sensitivity to noise as it removes the unconnected components of the segmentation. The final metric used was the average Hausdorff distance (AHD), which considers the mean of all the Hausdorff distances between the two segmentations. The AHD metric is less sensitive to outliers than the Hausdorff distance and provides an understanding of the magnitude of the distance between the segmentations that cannot be seen in the Dice coefficient. Using the Dice coefficient, Hausdorff distance, and AHD in conjunction with visual comparison provided an overall understanding of the differences in shape, size, outliers, and distance between the two compared segmentations (ground truth and algorithmic) while providing values that could easily be compared to previous segmentation projects in the literature [21].
Results
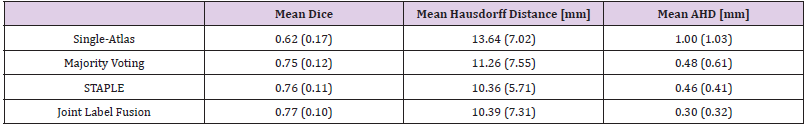
Results of the majority voting, STAPLE and joint label fusion metrics for segmentations created using just a single atlas (only one atlas is registered to the target image) as well as multi-atlas are shown in Table 1. For visual inspection, an example of an automated segmentation result in both two-dimensions (2D) and 3D is shown in Figure 3. All of the multi-atlas methods outperformed the singleatlas segmentations, which only produced a mean Dice of 0.62, a mean Hausdorff distance of 13.64 mm, and a mean AHD of 1.00 mm. Majority voting improved on single-atlas with a mean Dice of 0.75 with a mean AHD of 0.48 mm. STAPLE was found to give a mean Dice score of 0.76 with a standard deviation of 0.11. Joint label fusion’s resulting mean Dice score was slightly higher than STAPLE and provided a mean Dice of 0.77. Joint label fusion showed improvements in the average distance of 0.30 mm compared to STAPLE with an average distance of 0.46.
Table 1: Mean values of all metrics found from single atlas and multi-atlas approaches. Standard deviations given in parenthesis.
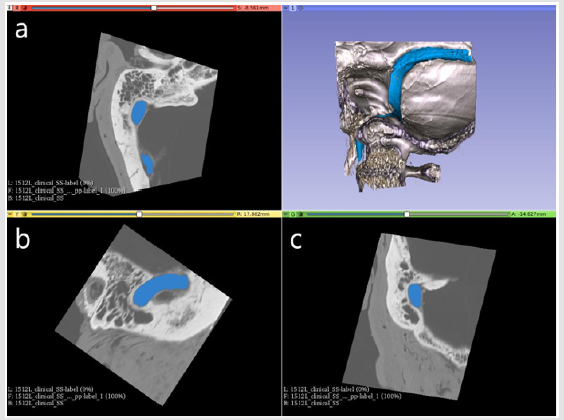
Figure 3: Screenshot from 3D Slicer [17] showing an example automated segmentation in three slice views (a axial, b sagittal, c coronal) as well as in a 3D rendered temporal bone segment. The automated segmentation and 3D model of the sigmoid sinus are shown in blue against the grayscale clinical-CT volume from which it was segmented.
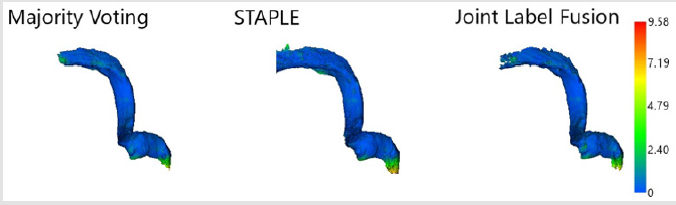
A comparison of the distances between the nearest points of the automated label fusion method results and the ground truth segmentation using absolute distance color maps can be seen in Figure 4. The colormaps revealed that the largest distance differences occurred at the inferior end near the jugular bulb within the jugular foramen where it connects to the jugular vein in the neck and the posterior extreme of the transverse portion of the sigmoid sinus of the segmented area of the sigmoid sinus. These areas are outside of the clinically relevant portion for surgical simulation. The distances were likely caused by differences in the size of the portion of the sigmoid sinus segmented between each ground truth model and the amount of the sigmoid sinus segmented by the algorithm.
Figure 4: Colormaps projected on automated segmentation 3D models showing absolute distance in millimeters between the algorithm segmentations and the ground truth sigmoid sinus.
Discussion
While the single-atlas approach to automated segmentation can occasionally provide comparable metric results to individual multi-atlas segmentations, the average performance of the singleatlas applied across the dataset of temporal bone images is much lower and the results are less consistent. This is likely due to the difficulty of capturing the high degree of variability of the sigmoid sinus anatomy in one example. This performance increase from single atlas to multi-atlas has been seen in previous works that segmented other anatomical structures, such as the brain from MRI scans [16,22]. The current methods presented for multiatlas- based segmentation of the sigmoid sinus provided accurate segmentations from clinical-CT scans which may be used in future surgical simulation. The use of joint label fusion, the most successful label fusion approach, resulted in a mean Dice coefficient score of 0.77, a mean Hausdorff distance of 10.39 mm, and a mean AHD of 0.30 mm, along with reasonable visual results.
The seemingly large mean Hausdorff distance may be attributed to the difference in the amount of the extremities segmented by the algorithm from the ground truth segmentations, since there were no large portions segmented outside the sigmoid sinus in the visual inspection. One drawback to joint label fusion is that the method requires much more computation time and storage space for registered images than STAPLE or majority voting, which performed almost as well as joint label fusion in the current paper. STAPLE and majority voting are therefore attractive options for segmenting the sigmoid sinus when time and storage space is at a premium. Additionally, while the present study focused on the structure of the sigmoid sinus, other structures of the temporal bone can be segmented using methods such as thresholding. Thresholding was not a viable option here due to the low contrast of the sigmoid sinus on the medial side.
By using μCT atlases, detailed, high-resolution models were created that had comparable metric scores to other temporal bone structures segmented by previous groups using different methods. Other atlas- based approaches for segmenting the structures of the temporal bone differ from the one presented herein, as they do not use multiple atlases for one segmentation, do not use label fusion methods to better capture variability, do not use μCT atlases, and do not target the sigmoid sinus [13,23]. Despite the accurate results produced by the present approach, it is important to note that since these are automatically generated models there is risk of error. If used clinically, automatically generated segmentations should be reviewed and revised as needed by an expert. Even in cases where the automated segmentation requires revision, automation significantly reduces the time and labor associated with manual segmentation.
Acknowledgment
This work was supported by the Otolaryngology Graduate Research Stipend from the Department of Otolaryngology - Head and Neck Surgery at Western University and by a Collaborative Health Research Project grant from the Canadian Institutes of Health Research and from the Natural Sciences and Engineering Research Council of Canada. Edits and review of the manuscript were performed by Lauren Siegel.
References
- A Arora, Khemani S, Tolley N, Singh A, Budge J, et al. (2012) Face and content validation of a virtual reality temporal bone simulator. Otolaryngol Head Neck Surg 146(3): 497-503.
- C Sewell, Morris D, Blevins NH, Dutta S, Agrawal S, et al. (2008) Providing metrics and performance feedback in a surgical simulator. Computer Aided Surgery 13(2): 63-81.
- D Morris, C Sewell, F Barbagli, K Salisbury, NH Blevins, et al. (2006) Visuohaptic simulation of bone surgery for training and evaluation. IEEE Comput Graph Appl 26(6): 48-57.
- GJ Wiet, Stredney D, Kerwin T, Hittle B, Fernandez SA, et al. (2012) Virtual temporal bone dissection system: OSU virtual temporal bone system: Development and Testing. Laryngoscope 122(SUPPL 1): S1-S12.
- R Varshney, Frenkiel S, Nguyen LH, Young M, Del Maestro, et al. (2014) The McGill simulator for endoscopic sinus surgery (MSESS): a validation study. J Otolaryngol Head Neck Surg 43(1): 40.
- B Tolsdorff, Pommert A, Höhne KH, Petersik A, Pflesser B, et al. (2010) Virtual reality: A new paranasal sinus surgery simulator. Laryngoscope 120(2): 420-426.
- MA Audette, H Delingette, A Fuchs, O Burgert, K Chinzei (2007) A topologically faithful, tissue- guided, spatially varying meshing strategy for computing patient-specific head models for endoscopic pituitary surgery simulation. In Computer Aided Surgery 12(1): 43-52.
- SM Anil, Y Kato, M Hayakawa, K Yoshida, S Nagahisha, et al. (2007) Virtual 3-dimensional preoperative planning with the dextroscope for excision of a 4th ventricular ependymoma. Minim Invasive Neurosurg 50(2): 65-70.
- S Weghorst, Airola C, Oppenheimer P, Edmond CV, Patience T, et al. (1998) Validation of the Madigan ESS simulator. Stud Health Technol Inform 50: 399-405.
- S Chan (2019) Cardinal Sim.
- R Jackler (2009) Atlas of skull base surgery and neurotology. Thieme: New York.
- K Van Osch, D Allen, B Gare, TJ Hudson, H Ladak, et al. (2019) Morphological analysis of sigmoid sinus anatomy: Clinical applications to neurotological surgery. J Otolaryngol Head Neck Surg 48(1): 2.
- KA Powell, T Liang, B Hittle, D Stredney, T Kerwin, et al. (2017) Atlas-Based Segmentation of Temporal Bone Anatomy. Int J Comput Assist Radiol Surg 12( 11): 1937-1944.
- H Wang, JW Suh, SR Das, JB Pluta, C Craige, et al. (2013) Multi-atlas segmentation with joint label fusion. IEEE Trans Pattern Anal Mach Intell 35(3): 611-623.
- P Aljabar, RA Heckemann, A Hammers, JV Hajnal, D Rueckert (2009) Multi-atlas-based segmentation of brain images: Atlas selection and its effect on accuracy. Neuroimage 46(3): 726-738.
- JE Iglesias, MR Sabuncu (2015) Multi-atlas segmentation of biomedical images: A survey. Med Image Anal 24(1): 205-219.
- A Fedorov, Beichel R, Kalpathy Cramer J, Finet J, Fillion Robin JC, et al. (2012) 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 30(9): 1323-1341.
- M Modat, DM Cash, P Daga, GP Winston, JS Duncan, et al. (2014) Global image registration using a symmetric block-matching approach. J Med imaging 1(2): 024003.
- M Modat, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, et al. (2010) Fast free-form deformation using graphics processing units. Comput Methods Programs Biomed 98(3): 278-284.
- SK Warfield, KH Zou, WM WellsMember (2004) Simultaneous Truth and Performance Level Estimation (STAPLE): An Algorithm for the Validation of Image Segmentation. IEEE Trans Med Imaging 23(7): 903-921.
- AA Taha, A Hanbury (2015) Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med Imaging 15(1): 29.
- RA Heckemann, JV Hajnal, P Aljabar, D Rueckert, A Hammers (2006) Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage 33(1): 115-126.
- JH Noble, BM Dawant, FM Warren, RF Labadie (2009) Automatic identification and 3D rendering of temporal bone anatomy. Oto Neurotol 30(4): 436-442.

 Research Article
Research Article