Short Communication
Application of Transcriptomic Biomarkers in
Livestock Product Safety
Xinyong You1,2, Qiong Li1 and Yali Zhou1*
Author Affiliations
1School of Biotechnology and Food Engineering, Anyang Institute of Technology, Anyang, China
2Henan Joint International Research Laboratory of Veterinary Biologics Research and Application, Anyang Institute of
Technology, Anyang, China
Received: August 07, 2019 | Published: August 19, 2019
Corresponding author: Yali Zhou, School of Biotechnology and Food Engineering, Anyang Institute of Technology, Anyang, China
DOI: 10.26717/BJSTR.2019.20.003486
In the research of livestock product quality and safety, the keys are to analyze, screen
and confirm illegal additives or contaminants added to livestock products. Searching
for biomarkers based on transcriptomics and using them to analyze the physiological
changes of animal tissues at the molecular level are becoming a novel and effective
approach for monitoring illegal use of contaminants in livestock products. This review
included the methods of studying transcriptomic biomarkers and latest progress of
applying the biomarkers to livestock product safety detection, aiming at offering new
technical measures to screen and detect additives or contaminants illegally added into
livestock products so as to guarantee the safety of livestock products.
Keywords: Transcriptomics; Biomarker; Livestock Product; Safety
Abbreviations: PCA: Principal Component Analysis; HCA: Hierarchical Cluster Analysis;
PLS-DA: Partial Least Squares Discriminant Analysis; GPs: Growth Promoters; BPSCs:
Bovine Prostatic Stromal Cells; TBA: Trenbolone Acetate
The application of illegal additives or contaminants in animal
breeding for livestock products severely impacts the development
of the livestock industry. Therefore, contaminant analysis and
screening become a hot topic among a multitude of researchers
at varied levels in different countries. Transcriptomics studies the
occurrence and change rules of the transcriptome in biological cells
based on the analysis of RNA-level gene expressions. It plays an
increasingly important role in studying the physiological activities
of tissues or cells and identifying the associations between gene
expressions and biological phenomena [1,2]. Biomarkers are
quantifiable biological molecules and represent the characteristics
of specific physiological states such as in a drug-interfered, normal
or pathological process, reflecting the molecular changes caused
by interactions between organisms and environmental factors. By
identifying and monitoring biomarkers, current biological processes
could be diagnosed and monitored in a timely manner. According
to previous researches, biomarkers have been successfully applied
to molecular medicine, medical diagnosis, disease predication,
risk evaluation, food safety, and so on. Identification of biomarkers
could be conducted at different molecular levels, such as the
genome, epigenome, transcriptome, proteome, metabolome and
liposome. Search for biomarkers based on -omics technologies and
quantitative determination of contaminants have become one of
food safety research focuses in recent years [3,4]. Researchers all
over the world have carried out biomarker researches targeting
at different contaminants in order to monitor the misuse of illegal
additives during livestock animal breeding.
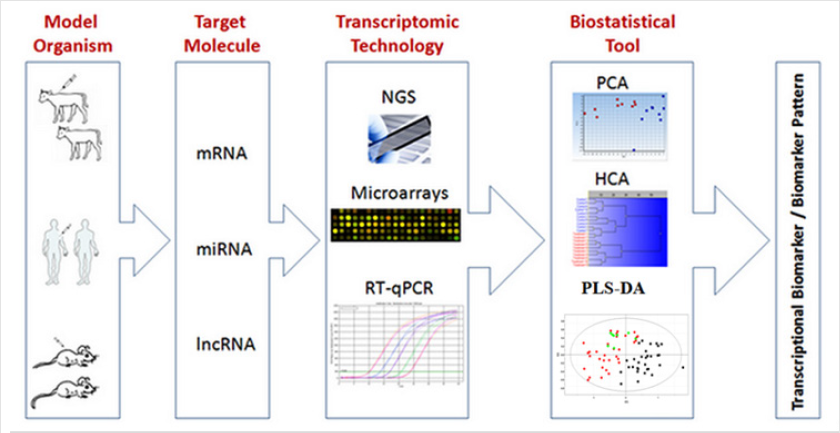
Transcriptional biomarkers are determined on the transcriptomic
platform using large-scale gene expression profiles, qRT-PCR
and biostatistical methods such as principal component analysis
(PCA), hierarchical cluster analysis (HCA) and partial least squares
discriminant analysis (PLS-DA). A standard route for finding and
developing gene expression biomarkers is as follows:
a. Setting up a high-quality sample set;
b. Using a large-scale gene expression platform;
c. Analyzing and identifying gene expression biomarkers by
mathematics and calculation;
d. Verifying the capability of the gene expression biomarkers
in differentiating independent samples.
Figure 1 shows the workflow of studying transcriptional
biomarkers [5]. Identification of gene expression biomarkers in
recent years provided a technical platform for studying livestock
product safety at different levels (Figure 1).
During livestock breeding, although certain veterinary drugs
have been prohibited, new drugs with similar effects and unknown
functional mechanisms made drug identification difficult. In search
for alternative detection methods, tracking physiological changes
of the components in these drugs to find out biomarkers has been
a focus. Earlier researches have indicated that the use of illegal
growth promoters (GPs) will bring about physiological changes in
animals at the molecular level. By analyzing and monitoring gene
expression changes in transcriptome using technologies such as
gene expression profiles and high-throughput RNA sequencing,
target genes in GP-treated animals could be screened, which can
serve as a novel and effective screening tool for monitoring illegal
use of GPs in animal-sourced food [4,6]. Riedmaier used RNA
sequencing to analyze the effects of dimethyl ether acetic acid and
estradiol on gene expressions of bovine livers. The expressions of
40 candidate genes were verified by qRT-PCR. Among them, 20
genes were significantly expressed. Then, a biostatistical tool for
model identification was employed to analyze the candidate genes,
which successfully differentiated samples in the treatment and
control groups.
These candidate genes were verified on the wild boars and calves
treated by synthesized metabolic agents. Moreover, Riedmaier et al.
found common biomarkers in cardiac and liver tissues of cattle. They
also determined the candidate genes according to the functional
mechanisms of different drugs on these tissues and monitored
the mRNA expression levels using fluorescent quantitative PCR.
Dynamic PCA was utilized to identify mRNA biomarkers to separate
the treatment group from the control group. Therefore, using RNA
sequencing, quantitative fluorescent PCR and biostatistical analysis
to screen candidate biomarkers have a great potential in detecting
illegally synthesized metabolic agents [6,7]. Pegolo studied the
transcriptional biomarkers in beef cattle treated with cortical
hormone. According to the comprehensive analysis of the DNAmicroarray
data of bovine muscle tissues in the control group, the
tissues treated by various GPs and those of unknown commercial
cattle samples, 73 gene expression biomarkers were found to
achieve high-precision classification with the Matthew correlation
coefficient of 0.77, and the percentages of false positives and false
negatives were 5% and 6%, respectively.
Although subject to the animal breed, age and GP treatment, the
test results showed that a group of relatively fewer genes could distinguish
the control group and cortical hormone treatment group,
providing transcriptional biomarkers for effective monitoring of GP
misuse in the process of livestock production [8,9]. Maria et al. reported
that gene expressions triggered by 17β-estradiol in bovine
prostate could be used to detect abuse of GPs. Quantitative PCR
and immunohistochemical methods were employed to analyze the
17β-estradiol triggered expressions of genes such as associated receptors in bovine prostatic stromal cells (BPSCs) and prostate tissues,
respectively. It was found that 17β-estradiol caused significant
overexpression of progesterone receptors in both BPSCs and prostate
tissues. Gene overexpression could still be detected 15 days
after animals were treated by 17β-estradiol. However, if chemical
methods were used, residuals could be detected only several hours
after animals were treated, revealing the advantage of PCR assay
using 17β-estradiol [10]. Uslenghi et al. used qPCR to achieve detection
of 17β-estradiol in bull on the basis of progesterone receptors
[11]. Divari found that progesterone upregulation could function as
a tool for detecting estrogen illegally used in adult beef cattle.
The monitoring of gene regulation at the mRNA level could be
used to detect illegal use of synthesized metabolic steroid in meat
production. In addition, qPCR was employed to identify illegal use
of 17β-estradiol and dexamethasone based on absolute quantitative
determination of oxytocin precursor genes, establishing the
sensitivity and specificity of the screening method. This method
was a novel screening tool and could significantly increase the
success rate of investigating animal food safety [12,13]. Starvaggi’s
research found that the expression of calmodulin in bovine tissues
could be used for monitoring illegal use of androgen in beef
breeding [14]. Carraro et al. studied the gene expression profiles
of the skeletal muscle and liver of bull treated by steroid GPs.
By observing the gene expression profile differences and using
qRT-PCR, biomarkers could be developed to detect illegal use of
dexamethasone in beef cattle production [15]. Cannizzo et al. used
morphological check and a transcriptomic approach to identify
the use of prednisone in beef cattle and found that a set of clear
transcriptional characteristics also supported monitoring of illegal
use of drugs in animal husbandry based on biomarkers [16,17].
Elgendy et al. conducted transcriptomic analysis in bovine muscle
and liver using both trenbolone acetate (TBA) and 17β-estradiol
(REV), used PCA on the microarray data to identify new candidate
biomarkers, and verified the biomarkers using onsite samples,
which could well differentiate animals in the control and treated
groups [18]. Zhao et al. determined 17 candidate genes by RNA
sequencing and literature research. They also used a biostatistical
method to obtain six key genes, which could be used as potential
transcriptomic biomarkers to monitor abuse of β2-receptor drugs
in lamb breeding [19,20]. You et al. used transcriptomic sequencing
and RT-qPCR to screen candidate genes in amantadine-treated
chicken breast muscle and liver tissues and preliminarily verified
that the candidate genes could be used as biomarkers to monitor
illegal use of amantadine drugs in broiler breeding [21].
Searching for biomarkers based on transcriptomics provides a
new approach for identifying illegal additives in livestock products.
It has two advantages, which are effective identification of the same
type of drugs and gene overexpression detection in the case of a
drug residual detection failure by a chemical method. According to
the present researches, it is defective using a set of common biomarkers
for all types of drugs that are illegally used in animals. A
single technique cannot adequately resolve the complexity of the
biological system. In contrast, a method integrating various -omics
technologies and data could become a key in searching for ideal,
stable, long effective and sensitive biomarkers. It is extremely important
to use -omics technologies to establish a reliable, specific
and sensitive screening method for quick detection of contaminants
in livestock products. With gradual progress and development of
technologies, analysis of contaminants or illegal additives in livestock
products using biomarkers will become increasingly mature.
We are grateful for the support by PhD Research Foundation
Project of Anyang Institute of Technology (NO. BSJ2019008), and
Henan Joint International Research Laboratory of Veterinary
Biologics Research and Application.
- Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10(1): 57-63.
- Lockhart DJ, Winzeler EA (2000) Genomics, gene expression and DNA arrays. Nature 405(6788): 827-836.
- Riedmaier I, Becker C, Pfaffl MW, Meyer HHD (2009) The use of omic technologies for biomarker development to trace functions of anabolic agents. J Chromatogr A 1216(46): 8192-8199.
- Riedmaier I, Pfaffl MW, Meyer HHD (2012) The physiological way: Monitoring RNA expression changes as new approach to combat illegal growth promoter application. Drug Test Anal 4(S1): 70-74.
- Riedmaier I, Pfaffl MW (2013) Transcriptional biomarkers-high throughput screening, quantitative verification, and bioinformatical validation methods. Methods 59(1): 3-9.
- Riedmaier I, Benes V, Blake J, Bretschneider N, Zinser C, et al. (2012) RNA-Sequencing as useful screening tool in the combat against the misuse of anabolic agents. Anal Chem 84(15): 6863-6868.
- Riedmaier I, Spornraft M, Pfaffl MW (2014) Identification of a potential gene expression biomarker signature in bovine liver to detect the abuse of growth promoters. Food Addit Contam A 31(4): 641-649.
- Pegolo S, Cannizzo FT, Biolatti B, Castagnaro M, Bargelloni L (2014) Transcriptomic profiling as a screening tool to detect trenbolone treatment in beef cattle. Res Vet Sci 96(3): 472-481.
- Pegolo S, Di CB, Montesissa C, Cannizzo FT, Biolatti B, et al. (2015) Toxicogenomic markers for corticosteroid treatment in beef cattle: Integrated analysis of transcriptomic data. Food Chem Toxicol 77: 1-11.
- Maria RD, Divari S, Bollo E, Cannizzo FT, Biolatti B, et al. (2009) 17β-oestradiol-induced gene expression in cattle prostate: biomarkers to detect illegal use of growth promoters. Vet Rec 164(15): 459-464.
- Uslenghi F, Divari S, Cannizzo FT, Maria RD, Spada F, et al. (2013) Application of absolute qPCR as a screening method to detect illicit 17β-oestradiol administration in male cattle. Food Addit Contam A 30(2): 253-263.
- Divari S, Mulasso C, Uslenghi F, Cannizzo FT, Spada F, et al. (2011) Progesterone receptor up-regulation: a diagnostic tool for the illicit use of oestrogens in adult beef cattle. Food Addit Contam A 28(12): 1677-1686.
- Divari S, Pregel P, Cannizzo FT, Starvaggi CL, Brina N, et al. (2013) Oxytocin precursor gene expression in bovine skeletal muscle is regulated by 17β-oestradiol and dexamethasone. Food Chem 141(4): 4358-4366.
- Starvaggi CL, Biolatti B, Sereno A, Cannizzo FT (2015) Regucalcin expression as a diagnostic tool for the illicit use of steroids in veal calves. J Agr Food Chem 63(23): 5702-5706.
- Carraro L, Ferraresso S, Cardazzo B, Romualdi C, Montesissa C, et al. (2009) Expression profiling of skeletal muscle in young bulls treated with steroidal growth promoters. Physiol Genomics 38(2): 138-148.
- Cannizzo FT, Pegolo S, Starvaggi CL, Bargelloni L, Divari S, et al. (2013) Gene expression profiling of thymus in beef cattle treated with prednisolone. Res Vet Sci 95(2): 540-547.
- Cannizzo FT, Pegolo S, Pregel P, Manuali E, Salamida S, et al. (2016) Morphological examination and transcriptomic profiling to identify prednisolone treatment in beef cattle. J Agr Food Chem 64(44): 8435-8446.
- Elgendy R, Giantin M, Montesissa C, Dacasto M (2015) The transcriptome of muscle and liver is responding differently to a combined trenbolone acetate and estradiol implant in cattle. Steroids 106: 1-8.
- Zhao L, Yang S, Zhang Y, Zhang Y, Hou C, et al. (2016) New analytical tool for the detection of ractopamine abuse in goat skeletal muscle by potential gene expression biomarkers. J Agr Food Chem 64(8): 1861-1867.
- Zhao L, Yang S, Cheng Y, Hou C, You X, et al. (2017) Identification of transcriptional biomarkers by RNA-sequencing for improved detection of β2-agonists abuse in goat skeletal muscle. Plos One 12(7): e0181695.
- You X, Xu M, Li Q, Zhang K, Hao G, et al. (2019) Discovery of potential transcriptional biomarkers in broiler chicken for detection of amantadine abuse based on RNA sequencing technology. Food Addit Contam A 36(2): 254-269.

 Short Communication
Short Communication