Abstract
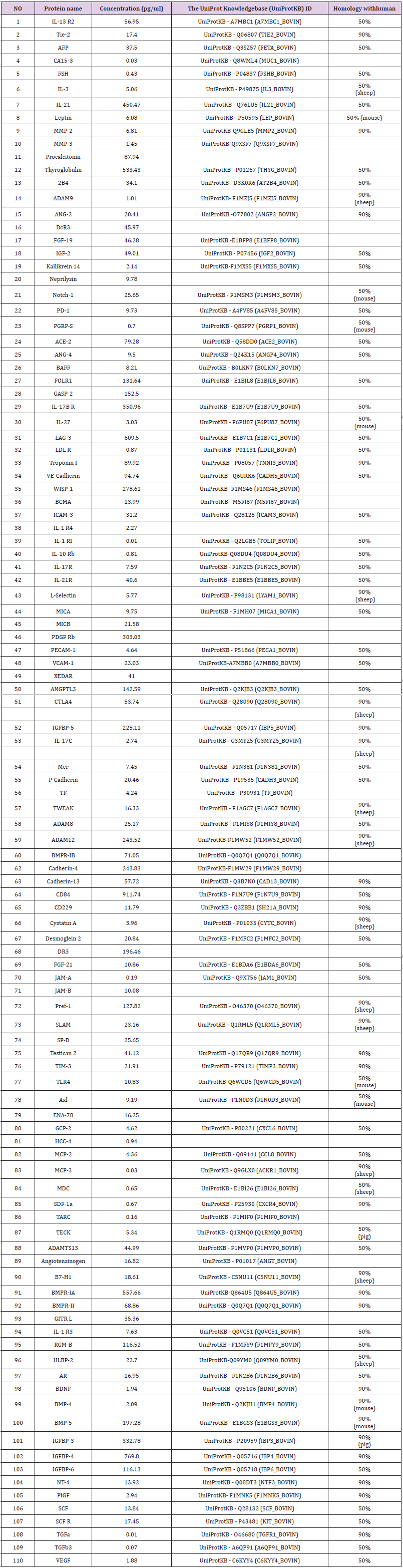
VitOrgan is a cell molecule drug extracted from bovine embryonic cells for anti-aging therapy since the 1965s in European countries. However, the molecular mechanisms involved remain unclear and underresearched. In this study, we applied human cytokine arraywith a group of 440 against-cytokine antibody, functionalGO term enrichmentand network technique to analyze the composition of vitOrgan(Mixed liquid: NeyPson Nr.5, NeyThel Nr.62, NeyNormin Nr.65, NeyDIL Nr.66, NeyDia Nr.67, NeyDIL Nr.70, NeyDesib Nr.78, NeyTroph Nr.96). The experimental results confirmed that the vitOrgan drug contains 123 cytokines, including ACE2, ADAM8, ADAM9, ADAMTS13, AFGF, ANGPT1, ANGPT2, ANG4, AR,AXL, BAFF,2B4,TNFRSF17, BDNF, bFGF, BMP4, BMP5, BMPR1A,BMPR1B, BMPR2,CDH4,CDH13, P-Cadherin, CD40L, CD84, CD229, CTLA4, CXCL9, CXCL10, CTA, TNFRSF6B, DCN, ENA-78,FGF19, FGF21, FOLR1, FSH, GPRASP1, GPRASP2, GCP-2, GITRL,HCC4,ICAM3,IFNA1,IFNB1,IFNG,IGF1,IGF2,IGFBP3,IGFBP4,IGFBP5,IGFBP6,IL1A, IL2, IL3, IL4, IL-10, IL-13, IL-15, IL-17A, IL-18, IL-21, IL-27, IL36RN, IL1RI, IL1RAP,IL1R4, IL10RB, IL13RA2, IL17RA, IL17BR, IL-17C, IL21R, F11R, JAM2, KLK14,LAG3, LDLR, LEP, LIF, CCL2, CCL8, CCL7, CL22, MER, MICA, MICB, CXCL9, CCL4, MMP-2, MMP-3, NACM1,MME,NOTCH1,NTF4,PDCD1, PECAM-1, PGYRP1,TGFB3,TG,TEK,HAVCR2, TLR4, TNFAIP3, TWEAK, VEGF, VCAM1, WISP-1, EDA2R and ULBP2.
Cluster analysis through biomedical informatics techniques suggestscytokines of vitOrganis a positive influences in cell biomedical regulation, including positive regulations of insulin-like growth factor receptor signaling pathway, cell proliferation, cell division, cell growth, ERK1 and ERK2 cascade, bone mineralization, epithelial cell proliferation, phosphatidylinositol 3-kinase signaling, protein kinase B signaling, glucose import, MAPK cascade, osteoblast differentiation pathway-restricted SMAD protein phosphoryl, cAMP metabolic process, tyrosine phosphorylation of Stat1 protein, participating immune response, proteoglycan biosynthetic process, inflammatory response, defense response to virus, chemokine-mediated signaling pathway, angiogenesis, growth factor activity, insulin secretion, DNA replication, lymphocyte chemotaxis, peptide catabolic process, endothelial cell apoptotic process and many other cell molecular activities. A preliminary interactome was built forthe bovine vitOrgan proteins. Our results confirm the great potential of the vitOrgan as a clinical relevant therapeutic strategy.
Introduction
The treatment known as live cell therapy was firstly reportedin Switzerland during the 1930s by Paul Niehans, which used organs, glands and fetuses of multiple species including sheep, cows and sharks [1,2]. In 1954, vitOrgan, freshlyextracted from bovine, was founded by Karl E. Theurer in German, which has been used as a targeting cell molecular drug for 65 years [3-5]. Since 1965,vitOrgan has been widely used in manyEuropean countries and become the first popular brand of anti-aging and functional sports medicine in Europe, which was reported several times [6,7]. Moreover, antiallergic therapy with vitOrgan is currently an effective method for hypersensitivityof immune system demonstrated by F.Heiss of the University Medical Center Hamburg-Eppendorf (UKE) [8].
F.Heiss used vitOrgan ALLERGOSTOP® as antiallergic agents in the clinical trials and obtained the follow-updata over a two-year period. In the study of pollen hypersensitivity, the ratioof the cured patients were 1224/1319(93%) for pollon hypersensitivity, and 491/593 (83%) for bronchial asthma, 681/783 (87%) forskin sensibility, and 183/382 (48%) for other allergies [8].
Rothschild also applies vitOrgan in the field of beauty and cosmetics, resulting in the birth of biolifting anti-aging therapy (https://vitorgan.de/). Neurodegenerative diseases are devastating and affect an estimated 1 billion individuals worldwide [9]. It is anticipated that cell-based drugs may alleviateor even reverse the progression ofneurological diseases[9]. In 2018 bovine myoblast cell productionin a microcarriers-based system was reported to provide valuable insights forclinical relevant cell therapy [10]. More than 40 years of clinical experience of vitOrgan has been accumulated targeting skin cosmetology, anti-aging, organ function maintenance, disease risk intervention, chronic disease adjuvant therapy and others. Cytokines are glycoproteins that help coordinate many physiological functions, including immune function, inflammation, hematopoiesis, homeostasis, and tissue repair [11,12].
Cytokine classification mainly include interferon (IFN-γ), interleukin (IL), colony stimulating factor (G-CSF) and erythropoietin (EPO) [11,13]. From 1982, the invention and productionof recombinant insulinto 1997 G-CSF became the first peptidedrug with annual sales exceeding 10 billion US dollars. Clinical indications of cytokines as supplementary therapy has relatively positive effects [14,15]. VitOrgan as live or fresh cell therapy is not only used in adjuvant treatment of daily diseases such as asthma, bronchitis, fatigue syndrome, sinusitis, immunodeficiency, diabetes mellitus, cardiovascular and cerebrovascular diseases, but also in the prevention and treatment of chronic motor diseases such as osteoarthritis, cartilage injury, intervertebral disc disease, etc. It is widely used in the field of anti-aging, preventative medication to reduce the risk of diseases. After treatment, the symptoms of many patients were significantly alleviated or even disappeared. Manypeople experience the improvement in the energy level, physical and sexual function, and organ function, which may indicate vitOrgan’s potential to delay the aging process [data under press]. Combining with the positive clinical outcomes from Germany, Brazil, Thailand, the Philippines, Austria, the United States, Russia, Colombia and othercountries, it is important to share the encouraging research data of vitOrgan with the research community. However, the molecular fraction and mechanism of vitOrgan is still unknown. In this study, we used cytokine microarray to detect the components of vitOrgan. Clustering and interaction analysis of vitOrgan-related cytokines were carried out using biomedical informatics technology through software in Database for Annotation, Visualization and Integrated Discovery (DAVID). These experimental data further providedmolecular basis for elucidating the mechanism of vitOrgan-related factorstherapy. Thus, our data would provide evidencefor cell and molecule therapy targetingtissue and organ to protect aging and damage.
Materials and Methods
Materials
The concentrated injection solution, vitOrgan of NeyPson Nr.5, NeyThel Nr.62, NeyNormin Nr.65, NeyDIL Nr.66, NeyDia Nr.67, were extracted from the bovine including Wagyu (black), Aberdeen Angus (black), Hereford (brown and white) and Brahman (grey and white).
Protein Array
We evaluatedthe quantitative concentrations of a total of 123 cytokines using the RayBio® G-Series Human Cytokine Antibody Array 440 (Cat# GSH-CAA-440, Array lot #Q0481618) according to the manufacturer’s instructions. Briefly, multiple cytokines specific capture antibodies were first bound to glass surfaces. After incubation with the plasma samples, the target cytokines were trapped on the solid surface as baits to capture the corresponding cytokines (or other mediators) in the applied vitOrgan samples (Mixed liquid: NeyPson Nr.5, NeyThel Nr.62, NeyNormin Nr.65, NeyDIL Nr.66, NeyDia Nr.67, NeyDIL Nr.70, NeyDesib Nr.78, NeyTroph Nr.96), and then incubated with a cocktail of prevalidated biotinylated secondary antibodies, and finally detected with Cy3-labeled streptavidin.
Gene Ontology and Pathway Analysis
Gene ontology (GO) and pathway analysis are suitable methods for integration genes with biological interaction and pathway networks to detect coordinated changes in functionally related genes. All the target genes are subjected to GO and pathway analysis in order to describe functional association of target genes. GO analysis was performed using GO pathway analysis using the open access software DAVID bioinformatics system and database (Database for Annotation, Visualization and Integrated Discovery, http://www.david.abcc.ncifcrf.gov website).Design and realization of software of cytokine statistical analysis for evaluation based on R language through KEGG (Kyoto Encyclopedia of Genes and Genomes https://www.genome.jp/kegg/) [16,17].
Data Analysis
Protein similarity analysis used the database of UniProt (https://www.uniprot.org/).A preliminary approach to the whole vitOrgan cytokines consisted of an elaboration for Network analysis with IPA software. As proteins in biological systems seldom work as independent entities, rather they are placed in widely interacting regulatory networks, the study of protein–protein interactions is a valid instrument to exploit in silico modelling to retrieve biologically relevant insights, through the individuation ofpivotal webs of proteins and their potential main modulators. Network analysis software, such as IPA, determines and graphs unbiased networks, in which gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least a reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base for IPA [16,17]. The databases and website including STRING (https://string-db.org/), National Center for Biotechnology Information (https://www.ncbi. nlm.nih.gov/) and GeneCardsSuite (https://www.genecards.org/).
Statistical Analyses
Data extraction can be done using the GAL file that is specific for this arrayalong with the microarray analysis software (GenePix, ScanArray Express,ArrayVision, MicroVigene, etc.).Comparison of means between two groups was accomplished by the Student’s t-test (two tailed). A P value <0.05 was considered statistically significant.
Results
Cytokine Expression Profiles
To identify the cytokines contained in vitOrgan injection solution for clinical applications, the cytokine profiles of vitOrgan samples (Mixed liquid: NeyPson Nr.5, NeyThel Nr.62, NeyNormin Nr.65, NeyDIL Nr.66, NeyDia Nr.67, NeyDIL Nr.70, NeyDesib Nr.78, NeyTroph Nr.96) were analyzed using a RayBio® G-Series Human Cytokine Antibody Array 440. The top ten cytokines contained in vitOrgan were CD84 (911.74pg/ml), LAG-3 (609.5pg/ml), BMPRIA( 557.66pg/ml), Thyroglobulin(533.43pg/ml), IL-21(450.47pg/ ml), IGFBP-4(769.80pg/ml), IL-17B R(350.96pg/ml), IGFBP- 3(332.78pg/ml), PDGF Rb (303.03pg/ml) and WISP-1(278.61pg/ ml) (Figure 1a & 1b), and CD84 was detected the highest level of the cytokine in samples (911.74pg/ml) (Figure 1).
We firstly used Human Cytokine Antibody Array to detect the molecular composition of the concentrated vitOrgan injection extracted from the bovine and found that Vitorgan contained the following 123 cytokines (Table 1). Among these 123 cytokines, 30 cytokineshadhigh concentrations (>100pg/ml), includingIL-13 R2, IL-21, IL-17B R, Procalcitonin, Thyroglobulin, ACE-2, FOLR1, GASP-2, LAG-3, Troponin I, VE-Cadherin, WISP-1, PDGF Rb, ANGPTL3, CTLA4, ADAM12, BMPR-IB, Cadherin-4, Cadherin-13, CD84, DR3, Pref-1, BMPR-IA, BMPR-II, RGM-B, BMP-5, IGFBP-5, IGFBP-3, IGFBP-4 and IGFBP-6 in the samples (Table 1). There were 79 cytokines atmoderate concentrations (100pg/ml-1 pg/ml), including Tie-2, AFP, IL-3, Leptin, MMP-2, MMP-3, 2B4, ADAM9, ANG-2, DcR, FGF-19, IGF-2, Kallikrein 14, Neprilysin, Notch-1, PD- 1, ANG-4, BAFF, IL-27, BCMA, ICAM-3, IL-1 R4, IL-17R, IL-21R, L-Selectin, MICA, MICB, PECAM-1, VCAM-1, XEDAR, IL-17C, Mer, P-Cadherin, TF, TWEAK, ADAM8, CD229, Cystatin A, Desmoglein 2, FGF-21, JAM-B, SLAM, SP-D, Testican 2, TIM-3, TLR4, Axl, ENA- 78, GCP-2, MCP-2, TECK, ADAMTS13, Angiotensinogen, B7-H1, GITR L, IL-1 R3, ULBP-2, AR, BDNF, BMP-4, NT-4, PIGF, SCF, SCF R and VEGF in the exposed samples (Table 1). Finally, fourteen cytokines have low concentrations (≤1 pg/ml), including IL-1 RI, IL-10 Rb, CA15-3, FSH, PGRP-S, LDL-R, JAM-A, HCC-4, MCP-3, MDC, SDF-1a, TARC, TGFa and TGFb3 in the exposed samples (Table 1). Acomplete list of identified proteins withinternational protein index accession number and the percentage of resemblence with human cytokineswere summarized in Table 1. The array allowed us to conduct the expression profiling of hundreds of cytokines, chemokines, growth factors, proteases, soluble receptors and other proteins involved in the important signal pathways.
Thus, our results showed that the vitOrgan injection solution contained a large numberof moleculesinvolved in the proteoglycan biosynthetic process, inflammatory response and defense responses. Furthermore, we searched the protein database of UniProt and found of the most abundantmoleculeswere 50%-90% resembling with the corresponding human protein sequences. In addition, 24 molecules did not show homology with human sequences, including IL-3, Leptin, ADAM9, Notch-1, PGRP-S, IL-27, L-Selectin, CTLA4, IL-17C, TWEAK, ADAM12, Cystatin A, Pref-1, SLAM, TLR4, Axl, MCP-3, MDC, TECK, B7-H1, ULBP-2, BMP-4, BMP- 5 and IGFBP-3 in the protein database of UniProt. Welisted their highest homology with mouse, sheep and pig sequences (50%- 90%) instead (Table 1).
GO Annotation Analysis of vitOrgan Cytokines
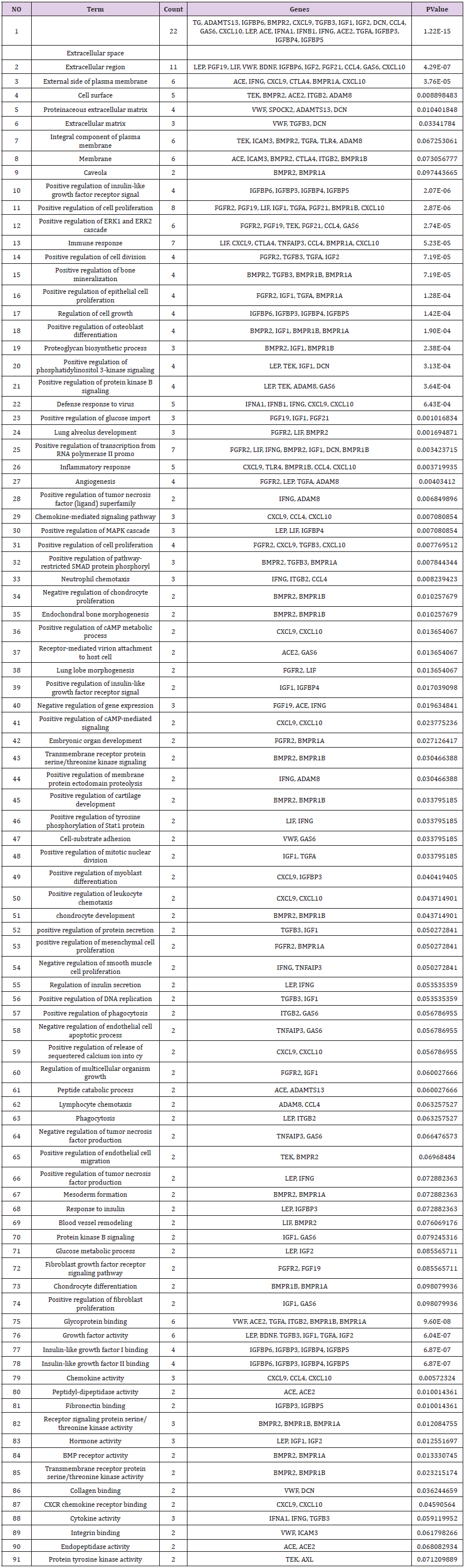
Network analysis through DAVID yielded the identification of 91 networks, as reported in Table 2. Most of these networks are accounted for biological functions related to extracellular space (22 cytokines), extracellular region (11 cytokines) and positive regulation of cell proliferation (8 cytokines). Networks involved in immune response were present as well (7 cytokines). The rest of the networks usually involve2-5 cytokines from vitOrgan. Most of the networks were intertwined, as they shared multiple entries. Indeed, certain categories of proteins were often recurring inmultiple networks, because of their multifaceted roles in cell activities and regulations, such as IGFBP3 (network 1, 10, 17, 49, 68, 77, 78 and 80), chemokines (network 1, 2, 3, 11, 13, 22, 26, 29, 31, 36, 41, 50, 59, 79 and 87) andgrowth factors (network 1, 2, 11, 12, 14, 16 and 23). The most significantly enriched pathways with the P-value are shown in Table 2.
Three outof the ten most abundant cytokines shown on Figure 1 participate in different signaling pathways inthe network analysis. For example, BMPR1A involved in 14 signal pathways usingnetwork analysis, including external side of plasma membrane, caveola, immune response, positive regulation of bone mineralization, positive regulation of epithelial cell proliferation, positive regulation of osteoblast differentiation, positive regulation of pathway-restricted SMAD protein phosphoryl, embryonic organ development, positive regulation of mesenchymal cell proliferation, mesoderm formation, chondrocyte differentiation, glycoprotein binding, receptor signaling protein serine/threonine kinase activity, BMP receptor activity (Table 2). IGFBP-3involved in 8 signal pathways by network analysis, includingextracellular space, positive regulation of insulin-like growth factor receptor signal, regulation of cell growth, positive regulation of myoblast differentiation, response to insulin, insulin-like growth factor I binding, insulin-like growth factor II binding, fibronectin binding (Table 2). IGFBP-4involved in 7 signal pathways by network analysis, includingextracellular space, positive regulation of insulin-like growth factor receptor signal, regulation of cell growth, positive regulation of MAPK cascade, positive regulation of insulinlike growth factor receptor signal, insulin-like growth factor I binding, insulin-like growth factor II binding.IGFBP-3 and IGFBP 4participated5signaling pathwaystogether, includingextracellular space, positive regulation of insulin-like growth factor receptor signal, regulation of cell growth, insulin-like growth factor I binding, insulin-like growth factor II binding (Table 2).
Figure 1: Cytokine antibody array of vitOrgan
A. Representative array images of top ten cytokine in vitOrgan. The 10 marker panel was annotated above and under the images.
B. Relative signal intensity of top ten cytokine contained in vitOrgan.
Statistical analysis was conducted on the enrichment of target genes that belong to each GO term and statistically significant values were calculated for each category. The significantly enriched GO terms were related to extracellular space (GO:0005615), external side of plasma membrane (GO:0009897), extracellular region (GO:0005576), cell surface (GO:0009986), proteinaceous extracellular matrix (GO:0005578), integral component of plasma membrane (GO:0005887), caveola (GO:0005901), immune response (GO:0006955), positive regulation of bone mineralization (GO:0030501), defense response to virus (GO:0051607), inflammatory response (GO:0006954), positive regulation of osteoblast differentiation (GO:0045669), chemokine-mediated signaling pathway (GO:0070098), positive regulation of pathwayrestricted SMAD protein phosphorylation (GO:0010862), positive regulation of phosphatidylinositol 3-kinase signaling (GO:0014068), positive regulation of protein kinase B signaling (GO:0051897), positive regulation of tumor necrosis factor (ligand)
superfamily member 11 production (GO:0000309), endochondral bone morphogenesis (GO:0060350), negative regulation of chondrocyte proliferation (GO:1902731), positive regulation of cell proliferation (GO:0008284), positive regulation of cAMP metabolic process (GO:0030816), positive regulation of transcription from RNA polymerase II promoter (GO:0045944), positive regulation of cAMP-mediated signaling (GO:0043950), proteoglycan biosynthetic process (GO:0030166), angiogenesis (GO:0001525), transmembrane receptor protein serine/threonine kinase signaling pathway (GO:0007178), positive regulation of membrane protein ectodomain proteolysis (GO:0051044), positive regulation of cartilage development (GO:0061036), positive regulation of tyrosine phosphorylation of Stat1 protein (GO:0042511), regulation of cell proliferation (GO:0042127), positive regulation of leukocyte chemotaxis (GO:0002690), chondrocyte development (GO:0002063), negative regulation of smooth muscle cell proliferation (GO:0048662), regulation of insulin secretion (GO:0050796), positive regulation of release of sequestered calcium ion into cytosol (GO:0051281), lung alveolus development (GO:0048286), lymphocyte chemotaxis (GO:0048247), positive regulation of endothelial cell migration (GO:0010595), positive regulation of tumor necrosis factor production (GO:0032760), mesoderm formation (GO:0001707), blood vessel remodeling (GO:0001974), positive regulation of cell division (GO:0051781), positive regulation of epithelial cell proliferation (GO:0050679), chondrocyte differentiation (GO:0002062), negative regulation of angiogenesis (GO:0016525), stem cell population maintenance (GO:0019827), positive regulation of MAPK cascade (GO:0043410), positive regulation of T cell proliferation (GO:0042102), positive regulation of peptidylserine phosphorylation of STAT protein (GO:0033141), neutrophil chemotaxis (GO:0030593), cellular response to lipopolysaccharide (GO:0071222), glycoprotein binding (GO:0001948), growth factor activity (GO:0008083), chemokine activity (GO:0008009), receptor signaling protein serine/threonine kinase activity (GO:0004702), BMP receptor activity (GO:0098821), transmembrane receptor protein serine/threonine kinase activity (GO:0004675), cytokine activity (GO:0005125), CXCR chemokine receptor binding (GO:0045236), and protein tyrosine kinase activity (GO:0004713), respectively (Figure 2).
The 20 most significantly enriched pathways with the significant P-value threshold of 10-4 were related to extracellular space (TG, ADAMTS13, IGFBP6, BMPR2, CXCL9, TGFB3, IGF1, IGF2, DCN, CCL4, GAS6, CXCL10, LEP, ACE, IFNA1, IFNB1, IFNG, ACE2, TGFA, IGFBP3, IGFBP4, IGFBP5), extracellular region (LEP, FGF19, LIF, VWF, BDNF, IGFBP6, IGF2, FGF21, CCL4, GAS6, CXCL10), external side of plasma membrane (ACE, IFNG, CXCL9, CTLA4, BMPR1A, CXCL10), positive regulation of insulin-like growth factor receptor signal (IGFBP6, IGFBP3, IGFBP4, IGFBP5), positive regulation of cell proliferation (FGFR2, FGF19, LIF, IGF1, TGFA, FGF21, BMPR1B, CXCL10), positive regulation of ERK1 and ERK2 cascade (FGFR2, FGF19, TEK, FGF21, CCL4, GAS6), immune response (LIF, CXCL9, CTLA4, TNFAIP3, CCL4, BMPR1A, CXCL10), positive regulation of cell division (FGFR2, TGFB3, TGFA, IGF2), positive regulation of bone mineralization (BMPR2, TGFB3, BMPR1B, BMPR1A), positive regulation of epithelial cell proliferation (FGFR2, IGF1, TGFA, BMPR1A), regulation of cell growth (IGFBP6, IGFBP3, IGFBP4, IGFBP5), positive regulation of osteoblast differentiation (BMPR2, IGF1, BMPR1B, BMPR1A),
proteoglycan biosynthetic process (BMPR2, IGF1, BMPR1B), positive regulation of phosphatidylinositol 3-kinase signaling (LEP, TEK, IGF1, DCN), positive regulation of protein kinase B signaling (LEP, TEK, ADAM8, GAS6), defense response to virus (IFNA1, IFNB1, IFNG, CXCL9, CXCL10), glycoprotein binding (VWF, ACE2, TGFA, ITGB2, BMPR1B, BMPR1A), growth factor activity (LEP, BDNF, TGFB3, IGF1, TGFA, IGF2), insulin-like growth factor I binding (IGFBP6, IGFBP3, IGFBP4, IGFBP5), insulin-like growth factor II binding (IGFBP6, IGFBP3, IGFBP4, IGFBP5) (Table 2).
Networks on the Whole Interactome of vitOrgan Cytokines
The networks of DAVID analysis on whole vitOrgan cytokines are shown in Figure 2. The thickness of the line represents the strength of the correlation, and the thicker lines demonstrated the stronger of the interaction. Networks with top score (Figure 3) included proteins involved in chemokine-mediated signaling pathway, immune response and inflammatory response et al. including IFN-β1, CCL8, TNFAIP3, CXCL6, CCL25, TNFSF12, TNFSF13B, CD40LG, PDCD1, IFN-α1, CCL4, CXCL9, CXCL5, CCL7, IL18, CXCL10, CCL5, IL15, IL2, IL3, CTLA4, CXCL12, IFN-γ, IL10, CCL2, IL17A, I L4, IL13, TLR4, IL1β, VEGFA, LEP, KITLG, FGF1, LIF, MMP3, CD36, MMP2, NOTCH1, FGF2, BDNF, PECAM1, TEK. The degree of a node indicates the number of links connected to a vertex. High degree nodes are the most relevant ones in interactome networks, as they represent the fulcrum of multiple signaling pathways, and their positive/negative modulation could result in alterations of the activities of their likely interactors. Sixteen main high-degree nodes were present in this network, namely CXCL9, CCL4, CXCL5, IL18, CXCL10, CCL5, IL17A, IL4, CCL2, IL10, IFN-γ, CXCL12, VEGFA, MMP2, FGF2, IL13, CTLA4, linked to proteins involved in chemokine-mediated signaling pathway and cell proliferation.
Figure 2: Gene ontology (GO) classification of cytokines in vitOrgan.
Gene ontology (GO) and KEGG pathway classification of target genes. The x-axis shows the GO categories and y-axis shows the target genes. The x-axis shows the P-value and y-axis shows the diverse biological functions of target genes according to the GO categories.
Figure 3: The network analysis on whole vitOrgan cytokines
Grey nodes, proteins from the dataset having a match within the database; white nodes, proteins from the database that were not identified (if present) upon the experimental phase; grey edges, interactions within a network; continuous line edge, direct interaction; interrupted line edge, indirect interaction
Vitorgan Cytokine-Cytokine Receptor Interaction
KEGG pathway is a collection of manually drawn pathway maps representing our knowledge on the molecular interaction, reaction and relation networks including metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, human diseases and drug development. Corresponding pathways can be predicted by KEGG analysis. Design and realization of software of vitOrgan cytokines analysis for evaluation is based on R Language and p value Cutoff = 0.05. We present a visualizing and integrating pathways using pathview package and demonstrated an interaction pathway which involved in as much more molecule of vitOrgan as possible.
This interaction showed 68 cytokines of vitOrgan and the red box labeled molecular denoted up-regulated cytokines, relating with chemokines (CCL25, CCL4,CCL4L1, CCL4L2, CCL17, CCL5, CCL8, CCL16, CCL7, CCL2, CCL12), CXC subfamily (CXCL5, CXCL6, CXCL9, CXCL10, CXCL12), the class I helical cytokines (IL-2, IL-4, IL15, IL3, IL4, IL13,IL27A, LIF), the class II helical cytokines(IL10, IL22, IFNA, IFNB1, IFNG), IL1-like cytokines(IL1B, IL1F5, IL18), IL17-like cytokines(IL17A, IL17B, IL17C, IL17RA), TNF family (DCR3, XEDAR, TWEAK, GITR, BCMA, CD40LG) and TGFβ family (TGFB3, BMPR2, BMPR1A, BMP4, BMPR1B, BMP5) (Figure 4). The results provide brief information on the possible mechanisms of cytokines from vitOrgan that are in effect for the therapy through data analysis.
Figure 4: The cytokine-cytokine receptor interaction of KEGG view on gene data with compound data on a global and overview map. Red box labeled molecular are related to up-regulated proteins, while green box labeled molecular are related to downregulated proteins.
Discussion
In this study, we focused onunderstanding the cytokine composition of concentrated vitOrganinjection solutionby using cytokine antibody array. As a result, a list of 123 molecules was found in the concentrated injection solution. To our knowledge, this is by far the most extensive inventory of molecules from vitOrganreportedinthe field. We have learned the types of molecules exist in vitOrgan and the types of cellular signaling pathways that these moleculesare intensively involved in.The results can help us anticipate the molecular mechanism related tothe treatment ofvitOrgan and can further provide theoretical guidance for potential clinical applications. On the other hand, these cytokines from vitOrgan may also participate in the repair and metabolism of organs. Potential targeted organs include:
(i) Thyroid(Thyroglobulin 533.43pg/ml present at top ten high concentrations in vitOrgan);
(ii) Brain (BDNF);
(iii) Bone (BMPRIA 557.66pg/ml, present at top ten high concentrations in vitOrgan; BMP-4, BMP-5, BMPRIB, BMPR2);
(iv) Reproductive tract (Testican 2) (Figure 1) and ( Table 1)[16]. Nevertheless, most cytokines of vitOrganhave been described acting on the whole body (Table 2), which suggest it may contribute to the homeostasis of the entire human physiological system.
The three molecules with the highest concentrationsamongcytokinesin vitOrganwere related in these 20most significantly enriched pathways including BMPRIA, IGFBP-3 and IGFBP-4. BMPRIA was involved in four most enriched pathways of external side of plasma membrane,immune response,positive regulation of bone mineralization and positive regulation of osteoblast differentiation (Figure 2). IGFBP-3 and IGFBP-4 together take part in four most enriched pathways of extracellular space, positive regulation of insulin- like growth factor receptor signal, regulation of cell growth and insulin-like growth factor I binding. In the case of IGFBPsit hasbeen suggested that they may regulate IGFs functions inpromotingcell proliferation/differentiation,because they:
L(i) Act as transport proteins in plasma and control theefflux of IGFs from the vascular space;
(ii) Prolong theIGFs half-lives and regulate their metabolic clearance;
(iii) Help coordinating tissue- and cell type-specific localizationof IGFs; and
(iv) Directly modulate interaction of IGFs withtheir receptors and thereby indirectly control its biologicalactions [16,18]. Therefore, the high concentrationscytokines were related to a series of cell and molecular functional activities and would play an important role in life and health maintenance. There are 93 molecules in vitOrgan resembles those in milk proteome compiling of 573 entries [16]. These moleculesare mainly involved in six network pathway, including
(i) Cell cycle, cancer, respiratory disease (VEGF, FGF-21, VEGFR2, EEF1G, EGF, bFGF, AFGF, FGF4, FGF7, FGF9, FGF17, FGFR2);
(ii) Lipid metabolism, molecular transport, small molecule biochemistry (TGFB1, TGFB2, TGFBI, TGFBR3, THBS1, LEP, BTC, EGFR, TGF-β2, IGFBP3, IGFBP1, IGFBP2, IGFBP4, IGFBP5, IGFBP6, IGFBP7);
(iii) Hematological system development and function, immune cell trafficking, inflammatory response (CXCL11, CXCL10, CXCL9, CCL4, ENA78, MMP-1, SDF-1, CX3CL1, CXCL11, CXCL12, CXCL13, CXCR3, GCP-2);
(iv) Carbohydrate metabolism, small molecule biochemistry, inflammatory response (IL-1 R3, CXCL1, IL-1R-1, IL-1R-2, IL1RL1, IL-2Rα, IL-5 Rα, IL-6R, IL-2RB, IL-lRa, CD44);
(v) Cellular development, cell death, hematological system development and function (GITR, IL-17C, IL-21R, TLR4, IFNβ, IL-1, IL-10, IFN-α, IFN-γ, IL-17A, IL-18, IL-2, IL-4, IL-15, IL-13, IL-1 f5, IL-21, IL-3, IL-5, IL-7, IL8, IL-9, IL-11, IL12B, IL16, IFN-γ, Fas(TNFRSF6), NGF-R, Osteoprotegerin, TRAIL R3, TRAIL R4);
(vi) Cardiovascular system development and function, cellular development, skeletaland muscular system development and function (PDGFRb, PDGF AA, PECAM-1, PDGF Rα, PDGF Rβ, TIMP-1, TIMP-2, TIMP3, PDGFB, PDGFRB) [16,19] (Figures 3 and 4). Milk is one of the physiological liquids with the most abundant nutrients. Many common cytokines shared between vitOragn and milk are related in various signaling pathways. Six pathways that shared ten cytokines between vitOrgan and bovine milk were listed above (i-vi) while the others with less shared cytokines were not listed. Because of such relevance, vitOrgan may help repair the damage to tissues and organs resulting from aging and environmental hazard, similar to the molecular mechanisms of bovine milk [16]. It would be helpful to study natural medicine in a scientific and systematic fashion to unveil the underlying mechanisms, which would point in the right development direction of natural medicine and complementary and alternative medicine in the future. Furthermore, the results of such a large biomedical data analysis require further clinical evaluation.
As we known live cell therapy(LCT)is an alternative treatment without medical evidence of effectiveness, which is through online marketing worldwide. It was reported the intramuscular injections of cell suspensions from fetal sheep,including young rams and pregnant ewes, were injected to human recipients for rejuvenation (anti-aging) and other ailments with positive feedback [20]. Apart from Germany recipients, medical tourists from North America and Asia travel to Germany to receive injections. Although several incidents were reported that LCT was the possible cause for the Q fever in Canada, Germany and the United States, vitOrgan have not experienced the issue. When working with vitOrgan, the health practitioners worldwide should pay more attention to the potential risk factors for LCT induced Q fever [21,22].
Altogether, this study focused on the cytokines involved in vitOrgan and is by far the first investigation about the molecular composition of vitOrgan injection solution. We have found 123 cytokines using human array and 91 important signal pathways associated with development, proliferation, differentiation, immunology and metabolism. Moreover, a preliminary map of vitOrgan proteins interactome was also constructed, which can greatly help understand the mechanism of vitOrgan. With more indepth knowledge of vitOrgan, clinician can put it into more targeted clinical applications and popularize the use of vitOrgan in the world.
References
- Robyn MP, Newman AP, Amato M, Walawander M, Kothe C, et al. (2015) Q Fever Outbreak Among Travelers to Germany Who Received Live Cell Therapy--United States and Canada, 2014. MMWR Morb Mortal Wkly Rep 64(38): 1071-1073.
- Rietschel HG (1953) Niehans fresh cell therapy. Medizinische 10(41): 1326-1329.
- Koch E (1955) Historical dates concerning Niehans fresh cell therapy and critical remarks on some priority claims. Hippokrates 26(20): 609-613.
- Pischinger A (1955) Fresh cell therapy; critical remarks on the theory and nature of Niehans' fresh cell therapy. Wien Med Wochenschr 105(46): 952-957.
- Jores A (1955) Critical comments on Niehans cellular therapy and on the methods of outsiders in medicine. Hippokrates 26(7): 206-209.
- Brauch F (1956) Fresh-cell therapy. Dtsch Med Wochenschr 81(27): 1086-1088.
- Bohl J, Goebel HH, Pötsch L, Esinger W, Walther G, et al. (1989) Complications following cell therapy. Z Rechtsmed 103(1):1-20.
- Heiss F (1991) Allergiebehandlung mit der Gegensensibilisierung(ALLERGOSTOP). Erfahrungsheikunde 40(8): 510-513.
- Dutta S, Singh G, Sreejith S, Mamidi MK, Husin JM, et al. (2013) Cell therapy: the final frontier for treatment of neurological diseases. CNS Neurosci Ther 19(1): 5-11.
- Verbruggen S, Luining D, van Essen A, Post MJ (2018) Bovine myoblast cell production in a microcarriers-based system. Cytotechnology 70(2): 503-512.
- Schooltink H, Rose John S (2002) Cytokines as therapeutic drugs. J Interferon Cytokine Res 22(5): 505-516.
- Azodi S, Jacobson S (2016) Cytokine Therapies in Neurological Disease. Neurotherapeutics 13(3): 555-561.
- Holdsworth SR, Gan PY (2015) Cytokines: Names and Numbers You Should Care About. Clin J Am Soc Nephrol 10(12): 2243-2254.
- Belardelli F, Ferrantini M (2002) Cytokines as a link between innate and adaptive antitumor immunity. Trends Immunol 23(4): 201-208.
- Olesen CM, Coskun M, Peyrin Biroulet L, Nielsen OH (2016) Mechanisms behind efficacy of tumor necrosis factor inhibitors in inflammatory bowel diseases. Pharmacol Ther 159: 110-119.
- D Alessandro A, Zolla L, Scaloni A (2011) The bovine milk proteome: cherishing, nourishing and fostering molecular complexity. An interactomics and functional overview. Mol Biosyst 7(3): 579-597.
- Shang Z, Li H (2017) Altered expression of four miRNA (miR-1238-3p, miR-202-3p, miR-630 and miR-766-3p) and their potential targets in peripheral blood from vitiligo patients. J Dermatol 44(10): 1138-1144.
- Rajaram S, Baylink DJ, Mohan S (1997) Insulin-like growth factor-binding proteins in serum and other biological fluids: regulation and functions. Endocr Rev 18(6): 801-831.
- Zhou J, Li F (2014) Potential pharmacokinetic interactions of therapeutic cytokines or cytokine modulators on small-molecule drugs: mechanistic understanding via studies using in vitro systems. Drug Metabol Drug Interact 29(1): 17-28.
- George M, Reich A, Cussler K, Jehl H, Burckhardt F (2017) Live Cell Therapy as Potential Risk Factor for Q Fever. Emerg Infect Dis 23(7): 1210-1212.
- Robyn MP, Newman AP, Amato M, Walawander M, Kothe C, et al. (2015) Q Fever Outbreak Among Travelers to Germany Who Received Live Cell Therapy-United States and Canada, 2014. MMWR Morb Mortal Wkly Rep 64(38): 1071-1073.
- Robyn MP, Newman AP, Amato M, Walawander M, Kothe C, et al. (2015) Q fever outbreak among travelers to Germany associated with live cell therapy - United States and Canada, 2014: a co-publication. Can Commun Dis Rep 41(10): 223-226.

 Research Article
Research Article