Short Communication
Computer Aided Screening of Indirubin Analogues
Targeting GSK-3β Protein Using Molecular Docking
Luu Van Huyen1, Tran Quoc Toan2, Pham Quoc Long2 and Pham Minh Quan2,3*
Author Affiliations
1Hanoi University of Natural Resources and Environment, Vietnam
2Institute of Natural Products Chemistry, Vietnam Academy of Science and Technology, Vietnam
3Graduate University of Science and Technology, Vietnam Academy of Science and Technology, Vietnam
Received: July 24, 2019 | Published: July 31, 2019
Corresponding author: Pham Minh Quan, Institute of Natural Products Chemistry, Vietnam Academy of Science and Technology,
Vietnam
DOI: 10.26717/BJSTR.2019.20.003414
Objective: To study the inhibition effects of indirubin analogues with GSK-3β protein
using molecular modeling and docking tools.
Materials and Methods: Crystal structure of GSK-3β was taken from Protein data bank
(PDB ID: 1Q41). The construction and optimization of the three-dimensional structure of
small molecules was done using ChemDraw and Chem3D version 18.1 (PerkinElmer®).
All ligands and protein were prepared for docking using Auto Dock Tools (ADT) 1.5.6.
AutoDock 4.2.6 was utilized for the molecular docking simulation.
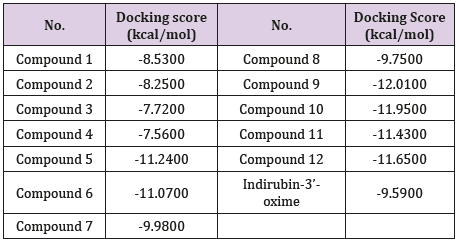
Results: Overall, amongst 12 indirubin derivatives, 8 analogues appear to inhibit
GSK-3β function based on docking score ranking, the formed hydrogen bonds of topmost
compounds share same residues in the binding site as compared to standard indirubin-
3’-oxime.
Conclusion: Further in vitro and in vivo experimentation are needed to confirm
the efficacy and potency of potential compounds. This model can be applied for design
candidate drug with desired biological properties by chemical modification in functional
group at appropriate places.
Keywords: Indirubin Derivatives; Docking; GSK-3β, Anti-Cancer
Abbreviations: PDB ID: Protein Data
Bank; ADT: Auto Dock Tools; NSCLC:
Non-Small-Cell Lung Cancer; SCLC:
Small Cell Lung Cancer
Lung cancer is one of the leading causes of cancer-related
death worldwide both with men and women, it is responsible for
about 1.3 million deaths annually [1,2]. In Vietnam, lung cancer
has very high rate of mortality with 24,000 new cases annually
in which 90% result in deaths. There are two major types of lung
cancer: non-small-cell lung cancer (NSCLC) and small cell lung
cancer (SCLC) [3,4]. Non-small cell lung cancer accounts for about
80% of overall lung cancer cases. Despite many efforts have been
made in the last 25 years, the treatment drugs for patients remain
poor efficacy. Indirubin and its analogues have been well known
for their activities against many types of cancer cell line, mainly
by inhibition of cell proliferation and induction of cell death [5,6].
These data suggest the development of indirubin derivatives
as potential candidates for the treatment of lung cancer. Recent
studies have discovered activity of GSK-3β in cancer and its role in
regulating tumor cell profileration and survival of multiple human
malignances. For the purpose of finding novel compounds and new
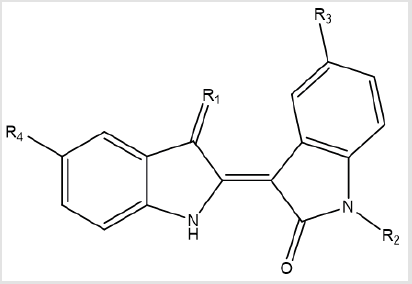
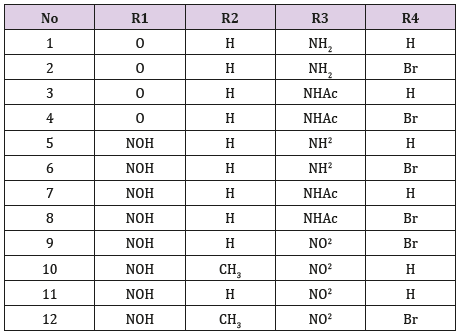
idea for drug design, 12 analogues of indirubin were allowed to
dock with GSK-3β protein.
Crystal structure of GSK-3β was taken from Protein data bank
(PDB ID: 1Q41). The construction and optimization of the threedimensional
structure of small molecules was done using Chem
Draw and Chem3D version 18.1 (PerkinElmer®). All ligands and
protein were prepared for docking using Auto Dock Tools (ADT)
1.5.6. The ligands and protein were protonated. The default Kollman
charges and solvation parameters were allocated to the protein
atoms. Gasteiger charges were added to each ligand atom. Grid
box was generated around possible ligand-binding site. The precalculated
binding affinity of each ligand’s atom type was prepared
using Autogrid. Auto Dock 4.2.6 was utilized for the molecular
docking simulation and Accelrys Discovery Studio Visualizer 4.0
was used for docking results visualization (Figure 1) and (Table 1).
In general, 8 derivatives of indirubin showed better docking
score to GSK-3β than the standard indirubin-3’-oxime. Most of the
designed derivatives form H-bonds with GSK-3β when binding in its
ATP-binding site. The hydrogen bonds occur mainly with VAL135
and ASP133 in the Hinge region and ARG14 which is the important
region for the function of protein. In some of the molecules it also
forms H-bonds with GLN185, LEU132, ASP200 and LYS88 amino
acids of GSK-3β (Table 2).
The authors declare that there is no conflict of interest.
Seung Hwa Hong, Kyu Sang Kyeong, Bang Yeon Hwang equally
contributed to this work.
- Fossella F, Pereira JR, von Pawel J, Pluzanska A, Gorbounova V, et al.
(2003) Randomized, multinational, phase III study of docetaxel plus
platinum combinations versus vinorelbine plus cisplatin for advanced
non-small-cell lung cancer: the TAX 326 study group. J Clin Oncol
21(16): 3016-3024.
- Biesalski HK, Bueno de Mesquita B, Chesson A, Chytil F, Grimble R, et al.
(1998) European Consensus Statement on Lung Cancer: risk factors and
prevention. Lung Cancer Panel. CA Cancer J Clin 48(3): 167-176.
- Shen YC, Chou CJ, Chiou WF, Chen CF (2001) Anti-inflammatory effects of
the partially purified extract of radix Stephaniae tetrandrae: comparative
studies of its active principles tetrandrine and fangchinoline on human
polymorphonuclear leukocyte functions. Mol Pharmacol 60(5): 1083-
1090.
- Choi HS, Kim HS, Min KR, Kim Y, Lim HK, et al. (2000) Anti-inflammatory
effects of fangchinoline and tetrandrine. J Ethnopharmacol 69(2): 173-
179.
- Eisenbrand G, Hippe F, Jakobs S, Muehlbeyer S (2004) Molecular
mechanisms of indirubin and its derivatives: novel anticancer molecules
with their origin in traditional Chinese phytomedicine. Journal of Cancer
Research and Clinical Oncology 130(11): 627-635.
- Karapetyan G, Chakrabarty K, Hein M, Langer P (2011) Synthesis
and bioactivity of carbohydrate derivatives of indigo, its isomers and
heteroanalogues. Chem Med Chem 6(1): 25-37.
Short Communication
Computer Aided Screening of Indirubin Analogues
Targeting GSK-3β Protein Using Molecular Docking
Luu Van Huyen1, Tran Quoc Toan2, Pham Quoc Long2 and Pham Minh Quan2,3*
Author Affiliations
1Hanoi University of Natural Resources and Environment, Vietnam
2Institute of Natural Products Chemistry, Vietnam Academy of Science and Technology, Vietnam
3Graduate University of Science and Technology, Vietnam Academy of Science and Technology, Vietnam
Received: July 24, 2019 | Published: July 31, 2019
Corresponding author: Young Chul Kim, Department of Physiology, Chungbuk National University, College of Medicine,
Chungdae-ro 1, Seowon-Gu, Cheongju 28644, Korea
Corresponding author: Pham Minh Quan, Institute of Natural Products Chemistry, Vietnam Academy of Science and Technology,
Vietnam
DOI: 10.26717/BJSTR.2019.20.003414
Objective: To study the inhibition effects of indirubin analogues with GSK-3β protein
using molecular modeling and docking tools.
Materials and Methods: Crystal structure of GSK-3β was taken from Protein data bank
(PDB ID: 1Q41). The construction and optimization of the three-dimensional structure of
small molecules was done using ChemDraw and Chem3D version 18.1 (PerkinElmer®).
All ligands and protein were prepared for docking using Auto Dock Tools (ADT) 1.5.6.
AutoDock 4.2.6 was utilized for the molecular docking simulation.
Results: Overall, amongst 12 indirubin derivatives, 8 analogues appear to inhibit
GSK-3β function based on docking score ranking, the formed hydrogen bonds of topmost
compounds share same residues in the binding site as compared to standard indirubin-
3’-oxime.
Conclusion: Further in vitro and in vivo experimentation are needed to confirm
the efficacy and potency of potential compounds. This model can be applied for design
candidate drug with desired biological properties by chemical modification in functional
group at appropriate places.
Keywords: Indirubin Derivatives; Docking; GSK-3β, Anti-Cancer
Abbreviations: PDB ID: Protein Data
Bank; ADT: Auto Dock Tools; NSCLC:
Non-Small-Cell Lung Cancer; SCLC:
Small Cell Lung Cancer

 Short Communication
Short Communication