Abstract
In previous study, we found that an oligodeoxynucleotide with CCT repeats (CCT ODN) could display immunosuppressive roles for TLR9 response in mice and in vitro. However, in this study, we found that CCT ODN could obviously elevate the mRNA levels of interferon-inducible protein 16 (IFI16) but no other DNA sensor such as AIM2, which belongs to same family of IFI16, in human monocyte line U937 cells when stimulated for 2 hours. At this condition, CCT ODN could also increase the level of interferon- (IFN-) mRNA in U937 cells. To confirm the CCT ODN-induced IFN- production associated with IFI16, we knocked down IFI16 expression and found that CCT ODN lost its induction role of IFN- production. Then, by using confocal microscope analysis, we found that IFI16 and CCT ODN could co-localize in the cytosol. The cytosol CCT ODN might be delivered by the cell surface DEC-205 that was down regulated after CCT ODN stimulation. Together, the study indicates that CCT ODN can activate IFI16 by associating with DEC-205 to mediate IFN- production in U937 cells, which may be the reason for explaining CCT ODN to attenuate the TLR9 response.
Abbreviations: SS DNA: Single-Stranded DNA; SLE: Systemic Lupus Erythematosus; STING: Stimulator of Interferon Genes; FBS: Fetal Bovine Serum; ATCC: American Type Culture Collection; SD: Standard Deviation; IRFs: Interferon Regulatory Factors
Keywords: SS DNA: Single-Stranded DNA; SLE: Systemic Lupus Erythematosus; STING: Stimulator of Interferon Genes; FBS: Fetal Bovine Serum; ATCC: American Type Culture Collection; SD: Standard Deviation; IRFs: Interferon Regulatory Factors
Introduction
The oligodeoxynucleotide with CCT repeats (CCT ODN) is a human microsatellite DNA sequence-based immunosuppressive ODN and displayed selectively negative regulatory role on TLR9 activation [1-3]. It can restrain TLR9 trafficking in immune cells, rescue mice from lethal shock sepsis induced by CpG ODN (TLR9 agonist) and decrease serum anti-DNA antibody levels in mice with lupus-like nephritis with unknown mechanism [4]. As an artificial synthesized single-stranded DNA (ssDNA), CCT ODN might display its role through certain DNA sensor. We know now, in addition to TLR9, there are several DNA sensors, such as DAI, RNA polymerase III, LRRFIP1, AIM2, IFI16 and others, which can sense intracellular DNA to initiate innate immune responses [5-8]. DAI, DNA-dependent activator of interferon regulatory factors (IRFs), is a first putative intracellular DNA sensor. It can bind double-stranded DNA (dsDNA) mimic poly(dA-dT) to induce IFN-α production in a very cell type specific [5]. RNA polymerase III also recognizes synthetic dsDNA-mimic poly(dA-dT). DNA-PKcs, Ku70 and Ku80 bind to DNA breaks when DNA damaged and promote DNA repair. LRRFIP1 (Leucine-rich repeat and flightless I interact protein 1) binds both DNA and RNA and induces IRF3 and NF-κB independent signaling pathway [6]. Both of IFI16 (interferon-inducible protein 16, also called AIM2-like protein) and AIM2 can sense dsDNA through their HIN domain in a sequence independent manner [7,8]. Among DNA sensors, IFI16 can also recognize single-stranded DNA (ssDNA) in a length dependent way and possibly acts as target for artificial synthesis ODN.
IFI16 has two HIN domains, HIN-A and HIN-B, which were separated by a spacer region. The HIN-B domain is predominant for IFI16. It was reported that IFI16 could induce ASC-dependent inflammasome activation during infection of nuclear DNA virus, such as HSV-1, KSHV and EBV [9-11] and act as a shuttle between cytosol and nucleus for DNA sensing. IFI16 could assemble on naked DNA strands based on length to distinguish self and non-self-DNA. Recently, there is a report that IFI16 is required for death of lymphoid CD4+ T cells abortively infected with HIV by holding up HIV DNA in cytosol, which results in caspase-1 activation and cell pyroptosis [12]. The author also confirmed that IFI16 is an only DNA sensor in charge of HIV DNA sensing, because knockdown another DNA sensor, including AIM2 and STING, cannot rescue HIV DNA-inducing cell death. It is interesting that AIM2 can also bind DNA in sequence-independent manner but cannot instead IFI16 in sensing HIV DNA. IFI16 is able to induce type-I IFN production and modulate a variety of cell functions including cell cycle, proliferation, differentiation, apoptosis/pyroptosis, senescence and inflammation [13-15], and also linked to the phathogenesis of certain autoimmune diseases, such as systemic lupus erythematosus (SLE) [16- 18]. STING (stimulator of interferon genes), as a mediator of type I IFN induction, can be recruited to IFI16 after IFI16 sensed DNA and activate IRF3 to drive the transcription of IFN and IFN-stimulated genes [19,20].
These studies hint IFI16 is a potential candidate for sensing ODN to mediate innate immune response. It is different from viral DNA produced in cells, ODNs contact intracellular DNA sensor only when they enter into cells. How the extracellular DNA is delivered into cells in which intracellular DNA sensor can sense it remains unclear. There is a report that DEC-205 could recognize extracellular CpG ODN and transport it into its target cells.DEC-205 is a professional endocytic receptor and plays an important role in antigen capture and presentation to T cells [21]. The structure property of DEC-205 suggests that it may potentially recognize multiple ligands and function as a “promiscuous” antigen receptor [22]. However, its natural ligands have yet not to be identified [23,24]. It calls attention to us to survey its possibility for extracellular CCT ODN transportation. In this study, we selected monocyte line U937 cells to be target cells for CCT ODN stimulation and detected IFI16 as necessary sensor of CCT ODN for cell response. The level of DEC-205 on the surface of U937 cells had been also observed after CCT ODN stimulation. Our results may provide a clue that extracellular DNA should be an activator for certain DNA sensor to initiate innate immune response, which may display anti-viral infection or neutralize a strong stimulation from other factors.
Materials and Methods
Cell Culture
U937 cells (human promyelocytic leukemia cell line, American Type Culture Collection (ATCC)) were obtained from Department of Molecular Biology, College of Basic Medical Sciences, Jilin University (Changchun, China). The cells were cultured at 37 °C in a 5% CO2 humidified incubator and maintained in RPMI 1640 medium supplemented with 10% (vol./vol.) heat-inactivated fetal bovine serum (FBS) and antibiotics (100 IU penicillin/ml and 100 IU streptomycin/ml). ODNs and Reagents The CCT ODN (5’-CCTCCTCCTCCTCCTCCTCCTCCT-3’) with full phosphorothioate modification was synthesized in Takara Co. (Dalian, China). It is diluted in PBS and tested for endotoxin by using the Limulus amebocyte lysate assay (Associates of Cape Cod, Inc.). All reagents used were pyrogen-free reagents. Pooled human AB sera were obtained from School of Stomatology, Jilin University (Changchun, China). IFI16 siRNA was purchased from Santa Cruz Biotechnology Inc. Rabbit anti-human DEC-205 andIFI16mAbs were purchased from MiltenyiBiotec (Auburn, CA). Goat antirabbit IgG FITC-labeled, was purchased from Zymed Laboratories (San Francisco, CA). Human IFN-α ELISA kit was purchased from eBioscience.
Flow Cytometry
To examine cell surface expression of DEC-205 proteins on U937 cells with or without CCT ODN stimulation, cells were plated into 24-well plates at a density of 1 × 106 cells/well. CCT ODN at 2 μM was added. After 2 hours’ incubating, cells were harvested and washed, then incubated for 30 min at 4 °C with primary anti-DEC-205 mAb containing 5% human AB serum. Unbound antibody was removed by washing the cells with PBS (4 °C). After centrifugation and two additional washing, the labeled cells were incubated at 37 °C for 30 min and stained with isotype-specific goat-anti-rabbit Alexa-488 secondary antibodies (Invitrogen). Then, the cell-associated immunofluorescence was measured by flow cytometry (FACScan; BD Biosciences).
Confocal Microscopy
For confocal imaging, U937 cells were allowed to adhere to poly L-lysine-coated cover slips, fixed in 4% paraformaldehyde in PBS and permeabilized with 0.1% triton-X100. After fixation and permeabilization, the cells were blocked in PBS containing 5% BSA and then stained with rabbit polyclonalprimary IgG overnight at 4 °C. Unbound antibodies were washed away and cells were incubated at 37 °C for 1 hour. Then, the cells were stained with isotype-specific goat anti-rabbit Alexa-488 secondary antibodies (Invitrogen), mounted with Mowiol (BrunschwigChemie, Amsterdam, The Netherlands) and imaged with a Bio-Rad MRC 1024 confocal system operating on a Nikon Optiphot microscope and a Nikon 100×planApo 1.4 oil immersion lens. Pictures were analyzed with BioRad-Lasersharp 2000 and Adobe Photoshop 7.0 software (Adobe Systems,Mountain View, CA, USA).
RNA Isolation and Quantitative RT-PCR
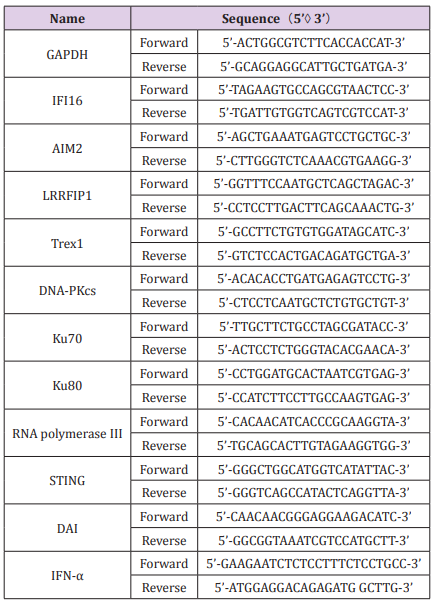
Total RNA was extracted from U937 cells using RNeasy Mini kit (Qiagen, Valencia, CA, USA) and reverse transcribed into cDNA using Taqman reverse transcription reagents (Applied Biosystems) with random hexamer primer, according to the manufacturer’s instructions. Real-time qPCR was performed using SYBR Green and LightCycler 480 system (Roche). The two-temperature cycle of 95 °C for 15 s and 60 °C for 1 min (repeated for 40 cycles) was used. Relative quantities of transcripts were calculated using the ΔΔCt method with GAPDH as a reference. The primers used for qPCR are synthesized by Sangon Biotech (Shanghai, China) and listed in Table 1.
Gel Electrophoresis and Western Blot
Whole cell extracts were prepared by washing cells twice with PBS(4 °C) and incubating them in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1%NP-40) with addition of phosphatase inhibitor cocktails 1 and 2 (Sigma; P2850 and P5726) and protease inhibitors (30 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, 10 g/ml aprotinin, 10 g/ml leupeptin, 1 g/ml pepstatin, 1 mM benzamidine) for 30 min at 4 °C on arocking platform before being scraped and transferred to tubes. The lysates were 103 PD performed with a DC protein assay kit (BioRad Laboratories). For Western blot analysis, 15 μg total protein was separated using standard 15% SDS-polyacrylamide gels and transferred to polyvinylidenedifluoride (PVDF) membranes (Immobilon P; Millipore). All blots were blocked in 5% skim milk in Tris-buffered saline (TBS)-Tween (0.1%) atroom temperature for 2 h, then read with the VersaDoc image system (BioRad, Hercules, CA, USA). Rabbit anti-human GAPDH, IFI16, and IFN-α mAbs were purchased from MiltenyiBiotec (Auburn, CA). Goat anti-rabbit IgG was purchased from Zymed Laboratories (San Francisco, CA).
ELISA for IFN-α
IFN-α production in the culture supernatants was measured using an ELISA kit (eBioscience) according to the manufacturer’s protocol. The optical density was detected at a wavelength of 450 nm on a microplatereader (Wellscan MK3; Labsystems). A standard curve ranging from 7.8 to 500 pg/ml was constructed using serial dilutions of a human IFN-α standard provided with the kit. Samples with values greater than the negative control but less than 7.8 pg/ ml were assigned values based on the extrapolation of the standard curve.
Small Interfering RNA and Transfection
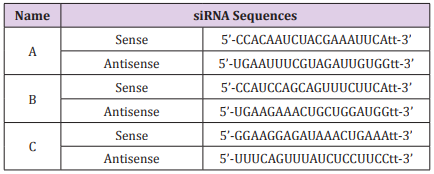
The siRNA oligonucleotides targeting IFI16 were purchased from Santa Cruz Biotechnology Inc. The IFI16-siRNA is a pool of three different siRNA duplexes of A, B and C (Table 2). A negative control siRNA 5’-r(UUCUCCGAACGUGUCACGU)d(TT)-3’ was used to monitor non-specific effects. 48 hours after transfection, Western blot was carried out to examine the knockdown of the targeted proteins.
Statistical Analysis
Results were representative of at least three independent experiments. The values were expressed as mean and standard deviation (SD) of individual samples. Statistical significance was determined by the Student’s t-test. A P<0.05 was considered a significant difference between the experimental groups.
Results
CCT ODN Induces the Expression of IFI16 and IFN-α in U937 Cells
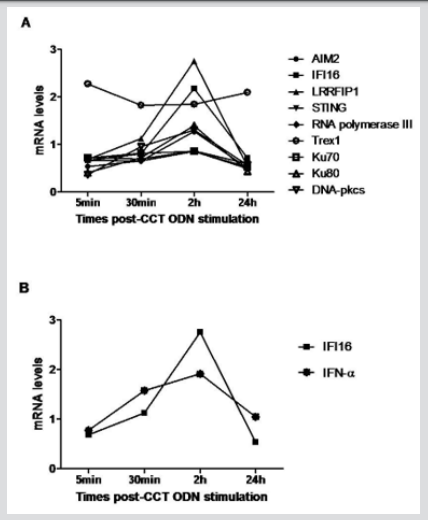
To observe whether CCT ODN triggering modulates DNA sensors in U937 cells, the expression of various known cytoplasmic DNA sensors was assessed by quantitative RT-PCR (qRT-PCR) after treatment with CCT ODN for different times. The result showed that CCT ODN could regulate the expression levels of different DNA sensor mRNA, including IFI16, LRRFIP1, Ku80, DNA-PKcs and AIM2, but not other DNA sensors when cultured the cells for 2 hours (Figure 1A). Among the up-regulated DNA sensors, IFI16 expression level was elevated at 30 minutes, reached peak at 2 hours and dropped down at 24 hours. When the cells treated by CCT ODN for 2 hours, the mRNA levels of IFI16 was reached to over 2.5-fold, which was higher over one-fold than that of AIM2, Ku80 and DNA-PKcs, and half fold than that of LRRFIP1. Because IFI16-STING pathway could induce type I interferon (IFN) production, we detected IFN-α mRNA in the cells by qRT-PCR. It was shown that CCT ODN could up-regulate IFN-α transcription in U937 cells after treated for 30 min and 2 hours. The peak of IFN-α mRNA was seen at 2-hour treatment of U937 cells with CCT ODN (Figure 1B). The results reveal that CCT ODN induces IFN-α production possibly through IFI16 signal pathway.
Figure 1: The effect of CCT ODN on the expression of DNA sensors and IFN-α in U937 cells. U937 cells were plated in 6-well plates at a density of 1x106,/ cells/well in RPMI 1640 medium/10% FCS, treated with CCT ODN (1 μM) for different times indicated in the figure, and then harvested for evaluating the mRNA levels of target genes by qRT-PCR. GAPDH mRNA level was as normalized standard. The data are representative of three independent experiments.
a) The levels of different DNA sensor mRNA.
b) Comparison of IFI16 and IFN-α mRNA levels.
IFI16 is Required for CCT ODN to INDUCE IFN-α Production in U937 Cells
To confirm the up-regulation role of CCT ODN for IFI16 expression, U937 cells were lysed after treatment with CCT ODN for 48 hours and the supernatant of the cell lysate was analyzed by Western blotting using anti-IFI16 antibody. The result revealed that there was certain level of IFI16 in U937 cells, which was upregulated by CCT ODN (Figure 2A). After assessed using grey values, it was found that CCT ODN could significantly increase the expression level of IFI16 in U937 cells (Figure 2B). To be clear the induction role of CCT ODN on IFN-α production was through IFI16 pathway, we firstly knocked down the IFI16 expression level using IFI16 siRNA (Figure 2A & 2B) and then detected the secretion levels of IFN-α from the cells. It was found that CCT ODN could significantly promote IFN-α secretion from U937 cells. The effect of CCT ODN could be abolished by IFI16 siRNA (Figure 2C), indicating IFI16 is required for CCT ODN inducing IFN-α production in U937 cells.
Figure 2: CCT ODN induces IFI16 and IFI16-dependent IFN-α production in U937 cells. U937 cells were cultured with CCT ODN directly or after IFI16 siRNA transfetion for 48 hours. The cells or cell supernatants were collected for western blotting with anti-IFI16 mAb or IFN-α detection by ELISA.
a) Western blot result of IFI16.
b) Grey values of the western blot results.
b) IFN-α levels in the supernatants of the U937 cells detected by ELISA. Data expressed as mean ± SEM
Co-Localization of CCT ODN with IFI16 in U937 Cells
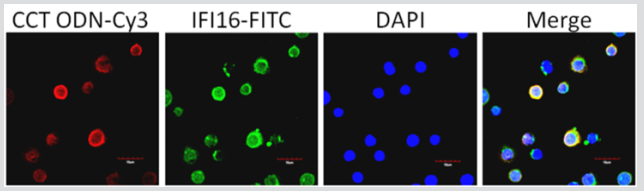
IFI16 as a DNA sensor has been shown to interact with various DNA localized in cytoplasm. To investigate whether IFI16 colocalizes with CCT ODN, we employed immunofluorescence confocal microscopy. The Figure 3 shows fluorescence images of U937 cells. The fluorescence of IFI16 (green) and CCT ODN (red) demonstrated a prominent cytoplasm punctate staining area. Merged images showed significant punctate areas of colocalization for IFI16 and CCT ODN in the cytoplasm of U937 cell. These data suggest that CCT ODN gets through the cell membrane into the cytoplasm and binds to IFI16.
DEC-205 as Surface Receptor for CCT ODN to Enter U937 Cells
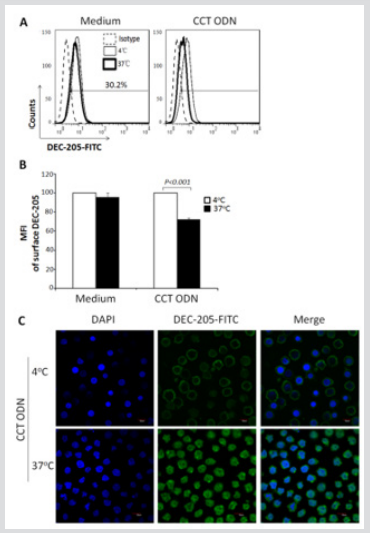
A recent report demonstrated that DEC-205, a C-type lectin, could act as a cell surface receptor for CpG ODN in human pDCs [25]. To survey the possible role of DEC-205 for delivering CCT ODN into U937 cells, the cells were labeled with anti-DEC-205 antibody followed by flow cytometry or confocal microscopy analysis. The results showed that 30.2 % of U937 cells were surface DEC-205 positive when cultured at 4 °C. At 4 °C culture condition, CCT ODN didn’t affect the level of DEC-205 on the cells. When the cells were cultured at 37 °C, CCT ODN decreased the level of DEC-205 on the surface of the cells (Figure 4A). By comparing the MFI of DEC-205 on the cells cultured either at 4 oC or at 37 oC, it was found that CCT ODN could significantly decrease the level of DEC-205 on the cells cultured at 37 °C (P<0.001) (Figure 4B), hinting DEC-205 may be internalized with CCT ODN together into the cells. To confirm the deduction, the stained cells were analyzed using confocal microscope. It was found that DEC-205 expressed on the surface of the cells when cultured at 4 oC, whereas moved into the cells when cultured at 37 °C (Figure 4C), indicating that DEC-205 can internalize into the cells along with CCT ODN.
Figure 3: Co-localization of CCT ODN and IFI16 in U937 cells. U937 cells were incubated with Cy3-labeled CCT ODN for 2 hours and then fixed on slides. The slides were stained with rabbit polyclonal IgG to human IFI16 overnight followed by addition of Alexa Fluor 488-conjugated goat anti-rabbit IgG for 1 hour at room temperature. All slides were visualized under a confocal microscope (x 1000). Data are representatives of three experiments.
Figure 4: Confocal image of DEC-205 on/in U937 cells. U937 cells were cultured with or without CCT ODN for 30 min at 4 °C or 37 °C and then stained or fixed followed by staining with anti-DEC-205 antibody. Afterward, the cells were analyzed by flow cytometry or a confocal microscope. Data are representatives of three independent experiments.
a) Data from flow cytometry analysis.
b) MFI of surface DEC-205 of U937 cells. Data are mean ± SEM of three independent experiments. Statistical significance determined by two-way ANOVA followed by Bonferroni test.
c) Confocal analysis of CCT ODN-activated U937 cells. Magnification: x 100.
Discussion
In this study, we showed that a human microsatellite DNA mimicking ODN with eight CCT repeats (CCT ODN) displayed an immune regulatory role in type I interferon induction in a human monocyte line U937 cells. This result is attractive to us because of its immunosuppressive activity showed in our previous research. To be clear the mechanism of CCT ODN, we tested its role of regulating intracellular DNA sensors. There is no report about regulatory ODNs (rODNs) plays their role through DNA sensor. As we known, rODNs can be divided into four types according their features of sequences and activities. Type I rODNs are guanine rich oligodeoxynucleotides, such as ODNs of 2114, 2088 and H154, and display inhibitory effect on CpG ODN-induced immune responses, such as proliferation of B cells, production of cytokines (e.g. IL-6, IL-12 and IFN-γ) by mouse spleen cells, secretion of IFN-α by pDCs, and so on. Type II rODNs are oligodeoxynucleotides with TTAGGG repeat based on mammalian chromosome telomere sequences, such as
A151. It is typical rODN with many immunosuppressive activities.
Usually, there are 500 - 3000 TTAGGG repeats in human chromosome telomeres. Type III rODNs are oligodeoxynucleotides with tandem quinine sequences and can inhibit its combination with CpG ODN or uptaking of CpG ODN. Type IV rODNs are longer sulfur modified oligodeoxynucleotides and display inhibitory roles by competing TLR9 with CpG ODN in a length-dependent manner. All of above rODNs are composed of G-rich sequences. However, CCT ODN presented in this study has no ‘G’ in its sequence and only contains CCT repeats, which are abundant in microsatellite DNA in human genome. In this point, CCT ODN should belong to a novel type of rODNs based on its immunosuppressive roles presented in our previous study but revealed stimulation activity on IFN-α induction through a cytosol DNA sensor IFI16, indicating any ODN may display negative role on one hand and positive role on the other hand. IFI16 can recognize both of ssDNA and dsDNA through its HIN domains. Although the length of naked ssDNA is thought to be necessary for IFI16’s DNA sensing, no one proved ODN could not target IFI16. In this study, we found CCT ODN could elevate the expression of IFI16 at mRNA and protein levels, which was connected to its induction activity of IFN-α production. This phenomenon hints IFI16 mediated pathway may attenuate TLR9-mediated pathway. TLR9 is for recognizing microorganism derived CpG DNA, whereas cytosol DNA sensor like IFI16 may be just for self-derived DNA to attenuate the strength of innate immune response through TLR pathway.It’s interesting that CCT ODN induces IFN-α production in U937 cells is IFI16 dependent. It is unknown how CCT ODN reached the IFI16, possibly by DEC-205 delivering. Why DEC-205, as surface receptor for CpG ODN, can distinguish CpG ODN and CCT ODN and send them to different cytosol receptor? So far, IFI16 is reported to recognize DNA in a sequence-independent manner. May be DEC205 binds different DNA by different motif, which lets IFI16 decide to sense or no-sense. All these questions need to be clarified in the future study. Anyway, our data in this article not only demonstrated IFI16, as a cytosolic DNA sensor, triggered type-I IFN pathway, but also revealed a novel mechanism of immune signaling in which CCT ODN was predominantly sensed by IFI16 along with DEC-205 internalization in U937 cells, and thereby triggered IFN-α production.
It remains to be determined whether other DNA sensors evolved in type I IFN pathway in the host as a mechanism of responding to CCT ODN. Identification of intracellular interaction partners of IFI16 will reveal how it modulates the type I IFN pathway.
Acknowledgement
We wish to thank the Department of Molecular Biology, College of Basic Medical Sciences, Jilin University (Changchun, China) for the cells.
References
- Hu D, Su X, Sun R, Yang G, Wang H, et al. (2009) Human microsatellite DNA mimicking oligodeoxynucleotides down-regulate TLR9-dependent and -independent activation of human immune cells. Molecular immunology 46: 1387-1396.
- Sun R, Sun L, Bao M, Zhang Y, Wang L, et al. (2010) A human microsatellite DNA-mimicking oligodeoxynucleotide with CCT repeats negatively regulates TLR7/9-mediated innate immune responses via selected TLR pathways. Clinical immunology 134(4): 262-76.
- He C, Zhou L, Sun R, Zhao T, Zhang Y, et al. (2011) Effects of oligodeoxynucleotide with CCT repeats on chronic graft versus host disease induced experimental lupus nephritis in mice. Clinical immunology 140(3): 300-306.
- Zhang X, Sun W, Wu X, Wang H, Yan Y, et al. (2014) An oligodeoxynucleotide with CCT repeats restrains CpG ODN-induced TLR9 trafficking. Current pharmaceutical biotechnology 15(9): 780-789.
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, et al. (2007) DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448(7152): 501-505.
- Yang P, An H, Liu X, Wen M, Zheng Y, et al. (2010) The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nature immunology 11(6): 487-494.
- Yan H, Dalal K, Hon BK, Youkharibache P, Lau D, et al. (2008) RPA nucleic acid-binding properties of IFI16-HIN200. Biochimica et biophysica acta 1784(7-8): 1087-1097.
- Jin T, Perry A, Jiang J, Smith P, Curry JA, et al. (2012) Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36(4): 561-571.
- Ansari MA, Singh VV, Dutta S, Veettil MV, Dutta D, et al. (2013) Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. Journal of virology 87(15): 8606-8623.
- Johnson KE, Chikoti L, Chandran B (2013) Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. Journal of virology 87(9): 5005-5018.
- Singh VV, Kerur N, Bottero V, Dutta S, Chakraborty S, et al. (2013) Kaposi's sarcoma-associated herpesvirus latency in endothelial and B cells activates gamma interferon-inducible protein 16-mediated inflammasomes. Journal of virology 87(8): 4417-4431.
- Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, et al. (2014) Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505(7484): 509-514.
- Gariglio M, Mondini M, De Andrea M, Landolfo S (2011) The multifaceted interferon-inducible p200 family proteins: from cell biology to human pathology. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research 31(1): 159-172.
- Gugliesi F, De Andrea M, Mondini M, Cappello P, Giovarelli M, et al. (2010) The proapoptotic activity of the Interferon-inducible gene IFI16 provides new insights into its etiopathogenetic role in autoimmunity. Journal of autoimmunity 35(2): 114-123.
- Duan X, Ponomareva L, Veeranki S, Panchanathan R, Dickerson E, et al. (2011) Differential roles for the interferon inducible IFI16 and AIM2 innate immune sensors for cytosolic DNA in cellular senescence of human fibroblasts. Molecular cancer research : MCR 9(5): 589-602.
- Costa S, Borgogna C, Mondini M, De Andrea M, Meroni PL, et al. (2011) Redistribution of the nuclear protein IFI16 into the cytoplasm of ultraviolet B-exposed keratinocytes as a mechanism of autoantigen processing. The British journal of dermatology 164(2): 282-290.
- Caneparo V, Cena T, De Andrea M, Dell'oste V, Stratta P, et al. (2013) Anti-IFI16 antibodies and their relation to disease characteristics in systemic lupus erythematosus. Lupus 22(6): 607-613.
- Gugliesi F, Bawadekar M, De Andrea M, Dell'Oste V, Caneparo V, et al. (2013) Nuclear DNA sensor IFI16 as circulating protein in autoimmune diseases is a signal of damage that impairs endothelial cells through high-affinity membrane binding. PloS One 8: 63045.
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, et al. (2010) IFI16 is an innate immune sensor for intracellular DNA. Nature immunology 11(11): 997-1004.
- Horan KA, Hansen K, Jakobsen MR, Holm CK, et al. (2013) Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. Journal of immunology 190(5): 2311-2319.
- Jiang W, Swiggard WJ, Heufler C, Peng M, Mirza A, et al. (1995) The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing. Nature 375: 151-155.
- Bonifaz L, Bonnyay D, Mahnke K, Rivera M, Nussenzweig MC, et al. (2002) Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. The Journal of experimental medicine 196(12): 1627-1638.
- Shrimpton RE, Butler M, Morel AS, Eren E, Hue SS, et al. (2009) CD205 (DEC-205): a recognition receptor for apoptotic and necrotic self. Molecular immunology 46: 1229-1239.
- Zhang SS, Park CG, Zhang P, Bartra SS, Plano GV, et al. (2008) Plasminogen activator Pla of Yersinia pestis utilizes murine DEC-205 (CD205) as a receptor to promote dissemination. The Journal of biological chemistry 283(46): 31511-31521.
- Lahoud MH, Ahmet F, Zhang JG, Meuter S, Policheni AN, et al. (2012) DEC-205 is a cell surface receptor for CpG oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America 109(4): 16270-16275.

 Research Article
Research Article