Abstract
The present work was carried out on the basis of the previously obtained results where a total of 71 accessions of the Cuban cocoyam collection of the genus Xanthosoma in Cuba were used, which were characterized through the 24 minimum descriptors selected among those that included the number of chromosomes and the color of rhizome flesh. The isoenzyme systems esterases, peroxidases and polyphenol oxidases were also analyzed. The isoenzimatic bands associated with the present cytotypes and the color of rhizome flesh were determined through the CHAID method (Chi-squared Automatic Interaction Detector). The isoenzymatic bands and association them with the chromosome numbers and the color of rhizome flesh were selected from the decision trees obtained with the CHAID method, which allowed to build classification rules (logical expressions). This method has already been used successfully in the characterization of risk factors and diagnosis of diseases by other authors, but in the available literature there were not found references on its application in plants.
From the practical point of view, having simple classification rules with high predictive value is very useful for in vitro early selection in genetic breeding programs, for in vitro conservation of germplasm to guide the collections of genetic material; in general, to detect early characteristics, such as the chromosome numbers and the color of rhizome flesh. They could also be used to detect interactions with predictive value, among other important characters on this and another genus. The created multiclassification rules allowed to achieve 100% of correct classification of the accessions according to the association of the isoenzymatic bands with the chromosome numbers and with the color of rhizome flesh, so they can be used as useful markers in the early determination of these characteristics.
Keywords: Variables; Classification; Accessions; Xanthosoma.spp
Introduction
In cocoyam, genus Xanthosoma, morphoagronomic data have been traditionally used for the characterization and identification of accessions [1] and they have been complemented with information on isoenzymatic markers and RAPD-type DNA polymorphism in integrated analysis, and 24 important minimum descriptors in the differentiation of cultivated accessions of this genus have been recommended recently (2 y 3); However, the possible association between some variables that allow the classification of accessions to be more efficient has not been determined, making use of the available characterization information. There is a variability that is detected with the naked eye and another that requires special techniques to be detected; for this reason, it is important to identify which of them is intended to be measured in order to be able to choose the appropriate statistical methods to analyze the data resulting from a characterization study. In general, publications on the subject contain little information about these methods and their use is not practical for most users. The main objective of this work is to determine the association between variables in the Cuban collection of cocoyam, genus Xanthosoma to classify the accessions and detect associated characteristics early with the use of statistical techniques.
Materials and Methods
The present work was carried out on the basis of the results obtained previously [2,3]. These authors used a total of 71 accessions from the Cuban cocoyam collection of the genus Xanthosoma in Cuba, which they characterized through the 24 minimum descriptors selected among those that included the number of chromosomes and the color of rhizome flesh. They also analyzed the isozyme systems esterases, peroxidases and polyphenoloxidases. The isoenzymatic esterase (Best)bands, peroxidase (Bprx) and polyphenoloxidase (Bppo) associated with the present cytotypes and the color of rhizome flesh were determined through the CHAID method (Chisquared Automatic Interaction Detector) of the statistical package SPSS® 15. The isoenzymatic bands and interactions between them with the chromosome numbers and the color of rhizome flesh were selected from the decision trees obtained with the CHAID method, which allowed to build classification rules (logical expressions).
Results
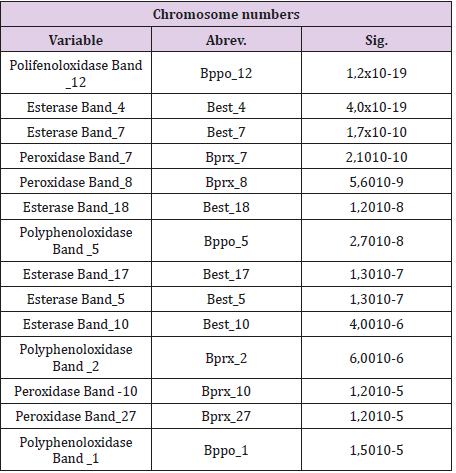
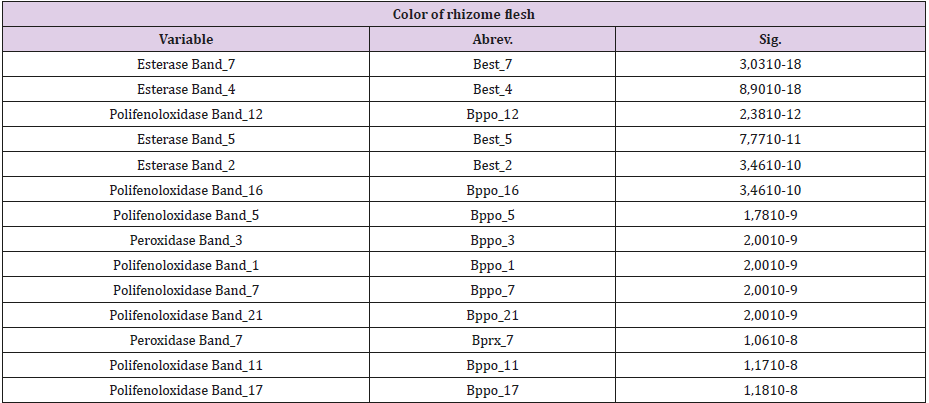
The CHAID technique allowed to determine the possible association of the isoenzymatic bands with the two cytotypes found and with the color of rhizome flesh of the accessions of the studied collection. This technique was able to segment the population into groups according to the presence (1) or absence (0) of the isoenzymatic bands and their interactions, which differentiate essential differences between the cytotypes and between the accessions with different color of rhizome flesh optimally. Using the CHAID method, it was also possible to detect the bands (of the three isoenzymatic systems studied) which have statistically significant associations with the two cytotypes found and with the color of rhizome flesh (Tables 1 & 3). A decision tree corresponds to each of these bands, in which the first segmentation of the population made at the root node is performed using this band (Figures 1-3).
Table 1: Isoenzymatic bands associated to the chromosome numbers in order of statistical significance of the variables.
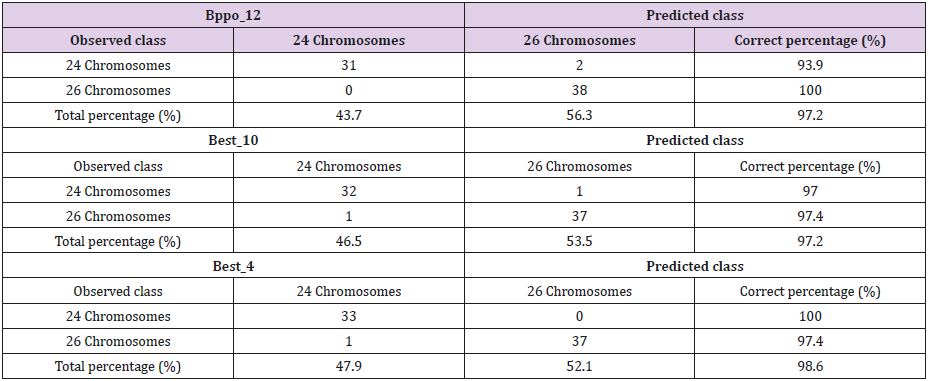
Table 2: Corresponding classification tables to the decision trees of the 71 accessions of the Cuban cocoyam collection, genus Xanthosoma studied, obtained through the CHAID method showing the association of the dependent variable, chromosome numbers and the predicted presence or absence of the most significant statistically bands of the three isoenzymatic systems studied.
Table 3: Isoenzymatic bands associated with the color of rhizome flesh in order of statistical significance of the variables.
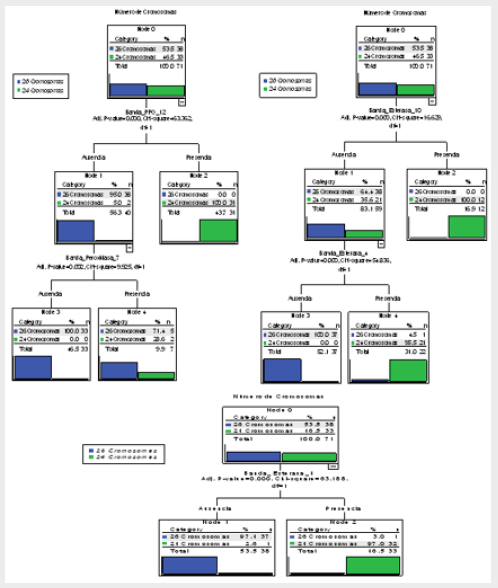
Figure 1: Decision Trees of the 71 accessions of the Cuban cocoyam collection, genus Xanthosoma studied, obtained through the CHAID method that shows the association of the dependent variable, chromosome numbers and the predicted presence or absence of the most significant statistically and most important biologically bands of the three isoenzymatic systems studied (Bppo_12, Best_10 and Best_4).
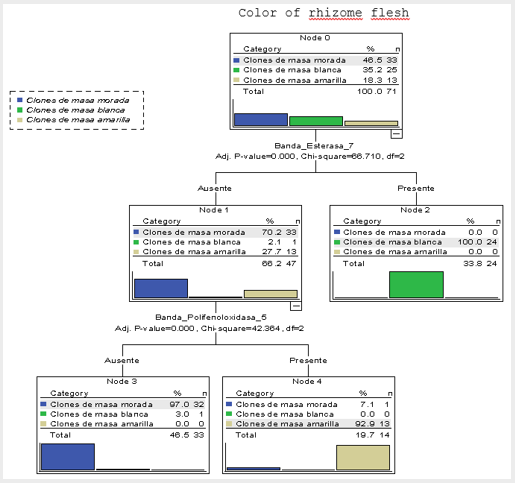
Figure 2: Decision tree of the 71 accessions of the Cuban cocoyam collection, genus Xanthosoma studied, obtained through the CHAID method that shows the association of the dependent variable, color of rhizome flesh and the predicted presence or absence of the most significant statistically bands (Best_7 and Bppo_5) of the three isoenzymatic systems studied.
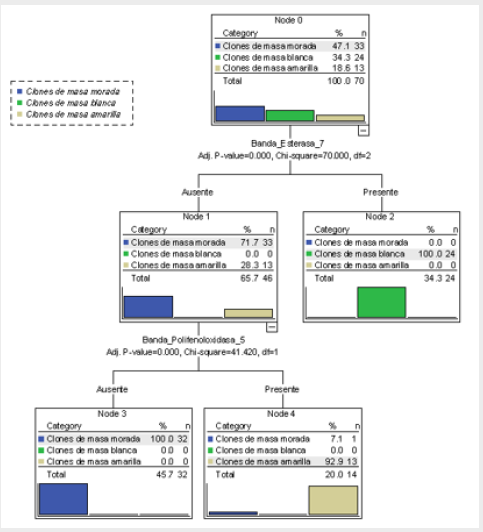
Figure 3: Decision tree of 70 accessions of the Cuban collection of cocoyam, genus Xanthosoma studied (the ‘Picante’ accession is excluded), obtained through the CHAID method that shows the association of the dependent variable, color of rhizome flesh and the predicted presence or absence of the most significant statistically bands of the three isoenzymatic systems studied (Best_7 and Bppo_5).
In order to build classification rules, the bands with the highest number of classified accessions correctly were selected from different decision trees. It should be pointed out that, in general, it is possible to build more than one classification rule for each cytotype and for each color of rhizome flesh. Through this work, the bands Best 4, Bprx_7, Bppo_12 and Best_10 were selected whose presence or absence can determine the existence of one or another cytogenetic type. The decision trees that involve these bands and their interactions are shown in Figure 1, while the classification derived from the corresponding decision trees is shown at Table 2. Although the classifications derived from each of the decision trees do not reach 100% of correct classification, a multiclassification rule, built from the main isoenzymatic bands, involved in these trees, achieves this objective. It was possible to develop rules that allow to classify 100% of the accessions correctly based on the chromosome numbers by knowing the association of the isoenzymatic bands (Table 1).
Next, the classification rules of the accessions with 2n = 24 and 2n = 26 chromosomes are exposed. An accession has 24 chromosomes if the following logical expression is true:
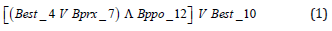
It is written literally:
If it is true that (Best_4 = 1 or Bprx_7 = 1) and Bppo_12 = 1) or if it is true that Best_10 = 1, then the accession has 24 chromosomes. On the other hand, an accession has 26 chromosomes if the following logical expression is true:
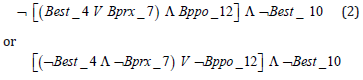
That is to say, if the given rule for n = 24 chromosomes is not met, then the denial of rule (1) is true1.In symbolic and simplified form it can be written: ¬ (1) = (2) or as in the previous case, it is written literally:
If it is true that (Best_4 = 0 or Bprx_7 = 0) and Bppo_12 = 0) or if it is true that Best_10 = 0, then the accession has 26 chromosomes. Then, the presence of Best_4, Bprx_7, Bppo_12 and Best_10 bands is associated in a biological and significant statistically way with accessions that have 2n = 24 chromosomes, and its absence is manifested in the accessions with a chromosome number of 2n = 26, so which it can be affirmed that these bands constitute isoenzymatic markers for the chromosome number in the studied collection (Figure 3).
An association between the chromosome number and flesh color is also observed, which is revealed in the presence of purple color in rhizome flesh of the accessions with 2n = 24 chromosomes. This fact constitutes an evidence in favor of the formulation of a hypothesis based on the Mendelian inheritance. The application of the CHAID method allowed the detection of several isoenzymatic markers and their interactions associated with the color of rhizome flesh. Table 3 shows the bands associated with the color of rhizome flesh, in a significant statistically way. The confirmation of these results makes possible to recommend these isoenzymatic bands as markers of use in future works of genetic breeding. Next, the classification rules of the accessions with white, purple or yellow color of rhizome flesh are shown. The accession produces rhizomes of white flesh, if the following logical expression is true:
Best _ 7 V Best _16 V Best _ 20 V Bprx _ 8 V Bprx _ 25 (3)
It is written literally:
If it is true that Best_7 = 1 or Best_16 = 1 or Best_20 = 1 or Bprx_8 = 1, then the accession produces rhizomes of white flesh.
The accession produces rhizomes of purple flesh, if the following logical expression is true:
¬Best _ 7 Λ (¬Bppo _ 5¬ Best _ 4) Λ¬Best _16 Λ¬Bprx _13 (4)
That is to say, if the following restrictions are met simultaneously:
1. Best_7 = 0
2. Bppo_5 = 0 or Best_4 = 1
3. Best_16 = 0
4. Bprx_13 = 0, then the accession produces rhizomes of purple flesh2.
It is written literally:
If it is true that Best_7 = 0 and (Bppo_5 = 0 or Best_4 = 1) and Best_16 = 0 and Bprx_13 = 0, then the accession produces rhizomes of purple flesh. The accession produces rhizomes of yellow flesh, if the following logical expression is true:
Bppo _16 V Bppo _17 V Best _ 2 (5)
It is written literally:
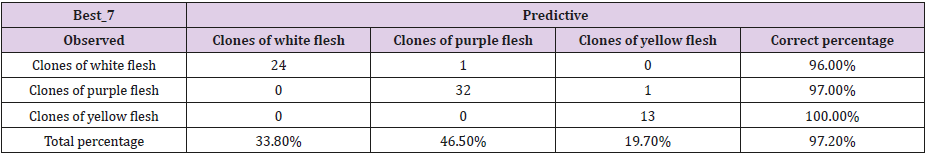
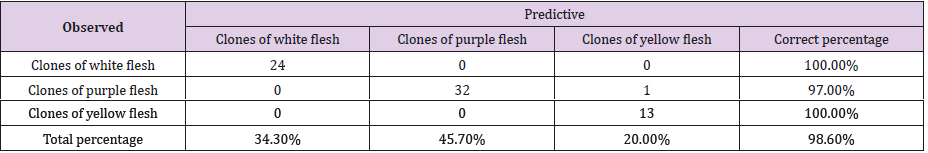
If it is true that Best_16 = 1 or Bppo_17 = 1 or Best_2 = 1, then the accession produces rhizomes of yellow flesh. The particularities of the ‘Picante’ accession which make it very different from the other accessions of the collection have been observed in all the analysis carried out through this work. In the decision trees of Figs. 2 and 3, and in the classification tables (Tables 4 & 5), it is observed how these results are confirmed; when this accession is included, the correct total percentage of classification is reduced from 98.6% to 97.2%.
Table 4: Corresponding classification to the decision tree of the 71 accessions of the Cuban collection of cocoyam, genus Xanthosoma studied, obtained through the CHAID method that shows the association of the dependent variable, color of rhizome flesh and the predicted presence or absence of the Best_7 band.
Table 5: Corresponding classification to the decision tree of 70 accessions of the Cuban collection of cocoyam, genus Xanthosoma studied (the ‘Picante’ accession is excluded) obtained through the CHAID method that shows the association of the dependent variable, color of rhizome flesh and the predicted presence or absence of Best_7 band.
The ‘Criolla1 1’ accession is not well classified by the decision trees obtained, which prevents reaching 100% of correct classification (Table 5). Although this accession has rhizomes of purple flesh and the rest of its morphoagronomic characteristics are similar to the accessions with that quality, its isoenzymatic patterns have a more similar behavior to the accessions with rhizomes of yellow flesh, hence the ambiguity offered by the decision tree in the classification of this accession. The previous results show the necessity and utility of applying multiclassifying rules, built with the main variables involved in the trees to reach 100% of correct classification [4].
From the practical point of view, having simple classification rules with high predictive value is very useful for in vitro early selection in genetic breeding programs, for in vitro conservation of germplasm to guide the collections of genetic material; in general, to detect early characteristics, such as the chromosome numbers and the color of rhizome flesh. They could also be used to detect interactions with predictive value, among other important characters on this and another genus. technique allows determining the possible association between variables. This technique is able to segment populations and detect their interactions to distinguish essential differences. The CHAID method has already been used successfully [5] in the characterization of risk factors and diagnosis of diseases, but in the available literature there were not found references on its application in plants.
Conclusion
The created multiclassification rules allowed to achieve 100% of correct classification of the accessions according to the association of the isoenzymatic bands with the chromosome numbers and with the color of rhizome flesh, so they can be used as useful markers in the early determination of these characteristics.
References
- MILIÁN M (2001) Caracterización de la variabilidad en la colección cubana de germoplasma de malanga Xanthosoma. Tesis de maestría pp. 130.
- MILIÁN M (2008) Caracterización de la variabilidad de los cultivares de la colección cubana de germoplasma del género Xanthosoma (Araceae). Tesis de Doctorado pp. 120.
- Milián M, O Molina Concepción, Y Figueroa Aguila (2018) Integrated Characterization of Cuban Germplasm of Cocoyam (Xanthosoma Sagittifolium (L.) Schott). Journal of Genetic Resources and Crop Evolution 1(1): 1-18.
- Franco T, E Hidalgo R (2003) Análisis estadístico de datos de caracterización morfoagronómica de recursos fitogenéticos. IPGRI. Boletín Técnico no. 8.
- Zamora L, R Grau, G Cardoso Y, M Alegret (1997) Una técnica de segmentación aplicada a la epidemiología y al diagnóstico. Tesis presentada en opción al título de Master en Matemática Aplicada. Universidad Central “Marta Abreu” de Las Villas, Santa Clara, Villa Clara, Cuba.

 Short Communication
Short Communication