Abstract
Epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs) are highly promising drugs that are well tolerated and have good antitumor activity in nonsmall cell lung cancer (NSCLC) patients with sensitive mutations in the EGFR gene. Here we reviewed 9 databases, including CKB, CIVIC, PMKB, COSMIC, My Cancer Genome, OncoKB, FDA, ClinicalTrials.gov and NCCN, in which we can obtain the EGFR mutants and related target drug annotations. Some of them have launched the information of EGFR TKIs and related activating EGFR mutations for advanced NSCLC, such as My Cancer Genome, CKB and CIVIC. Although these databases have overlaps in the variants and annotations, their major contents vary. This leads the increase of complexity of annotating target drug information for the EGFR mutants.
Keywords: Annotation; EGFR TKI; NSCLC
Abbreviations: ORR: Objective Response Rate; PFS: Progression-Free Survival; OS: Overall Survival
Introduction
Lung cancer has the highest incidence among all cancers and is the leading cause of cancer-related death worldwide. Non-small-cell lung cancer (NSCLC) accounts for approximately 80% of lung cancer cases [1]. In more than 40% NSCLC patients, the gene encoding epidermal growth factor receptor (EGFR), a transmembrane glycoprotein, is overexpressed and further mutant EGFR is present in more than 50% Asia NSCLC patients [2]. EGFR is a member closely related to growth factor receptor tyrosine kinases (RTKs) that consist of four members: EGFR (HER1/ErbB1), HER2 (ErbB2), HER3 (ErbB3) and HER4 (ErbB4), which regulate many developmental, metabolic and physiological processes [3]. Activating mutations within the tyrosine kinase domain of the EGFR are found in more than 40% of the Asia lung adenocarcinomas [4]. The EGFR-tyrosine kinase inhibitors (TKIs) are highly promising drugs that are well tolerated and have good antitumor activity in NSCLC patients with sensitive mutations in the EGFR gene [5]. In particular, the use of EGFR-TKIs in the treatment of advanced NSCLC patients harboring activating EGFR mutations has markedly improved survival outcomes, particularly in Asian descent patients [6]. While, the drugs resistance is the most challenging problem of the EGFR-TKI treatment in NSCLC patients. The major mechanisms giving rise to EGFR-TKI resistance in NSCLC have been demonstrated, including secondary mutation of EGFR T790M and other related positions [7].
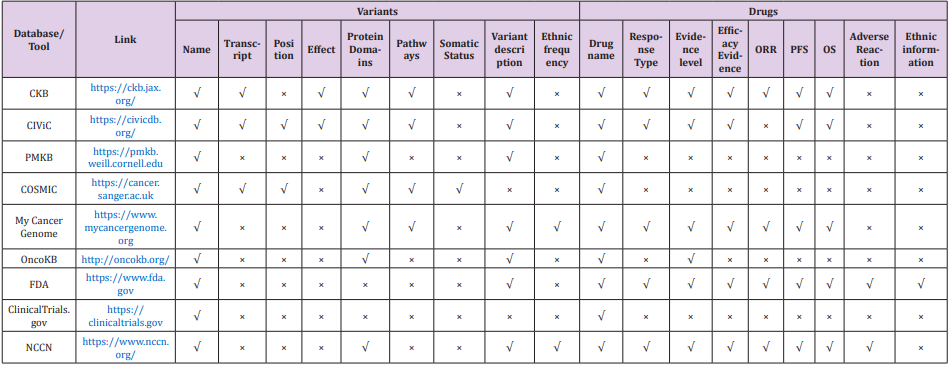
Due to the development of sequencing technology, it is getting easier and cheaper to identify mutations in a particular gene or in the genome. Many bioinformatics tools have been developed to characterize gene mutations, such as BWA [8,9], GATK [10] and VarScan [11], and to annotate the mutations, such as ANNOVAR [12] and SnpEff [13]. However, neither provides the target drug information related to gene variants, which is still not working sufficient in clinical sequencing workflow [14]. To obtain such information, users have to search in some other databases, such as MyCancerGenome [15], OncoKB [16], ClinicalTrials.gov [17], CIVIC [18] and PMKB [19]. Although all these databases provide clinical information of EGFR mutations and target drug information, they focus on different areas and a combination of these databases can help to understand the EGFR-TKI mutations and target drug information. In this review we summarized and compared 9 databases (Table 1) that can help annotate the EGFR-TKI mutations and related target drug information.
Comparison of 9 Public Databases
The major contents vary among the 9 databases (Table 1). Basic information of related drug (e.g., name) can be searched using the name of EGFR mutant in all databases. While, detailed information about the variants or the drugs are selected to present in the search results with different database (Table 2). Among them, CKB, CIVIC, PMKB, COSMIC and MyCancerGenome can provide some information of the EGFR mutant, such as the transcript, effect of the mutation, protein domain, pathways and ethnic frequency in population. While OncoKB, FDA, ClinicalTrials.gov and NCCN are more focusing the drugs. In the FDA website users can obtain the most detailed information about target drugs using the name of EFGR mutant.
Table 2: Detailed information about the variants or the drugs are selected to present in the search results with different database.
Note: Position refers to genomic coordinates, such as chr7:55249071.55249071.
Note: Effect refers to gain-of-function or loss-of-function.
Note: Variant description refers to the details, such as protein domain, biochemically, protein function, report information of the variant.
Note: Response Type refers to reaction to the drug, such as sensitive or resistant.
Note: Evidence level refers to reliability, such as FDA approved or clinical study and so on.
The characteristics of each database are as follows:
a. The main display information of CKB database is classified by three parts – gene variants, molecular profiles and gene level evidence. It provides information as comprehensive as possible and uses a clear information presentation way for users to understand. But complete information of EGFR is not directly available for free.
b. The CIVIC database provides information as comprehensive as possible, which includes much data from timely public reference papers. While the ethnic information is missing, and it takes time to filter the results from a lot of hits with low-level evidence.
c. The PMKB database is mainly classified by three parts – gene, variant and interpretation. However, no detailed drug related information can be found in this database, such as ORR, median PFS, median OS and adverse reaction.
d. The COSMIC database is a relative complete database for gene variants. It can be better if some more information of the drugs is provided, rather than the names only.
e. The MyCancerGenome database provides relative full information. The annotated information can be chosen by disease, gene and variant. But the information of drugs is not as detailed as the FDA.
f. The OncoKB database provides information classified by evidence level. It links the gene variants with related drugs. However, detailed information is missing for the variants and the drugs, such as effect for variants, ORR, median PFS, median OS and adverse reaction for drugs.
g. The FDA database releases comprehensive and authoritative for approved drugs, but no detail information for gene variants can be found, such as effect, protein domains and pathways.
h. The Clinical Trials database provides clinical trial information from most countries in the world.
i. The NCCN database releases guidelines for clinical practice of malignant tumors, which includes therapy choice, such as how to choose drugs according to situations. But detail information for gene variants is relatively lack, such as effect and pathways.
Conclusion
The current public target drug annotation databases all have advantages and disadvantages. When doing annotations for specific genetic variants, users need to integrate the information of them. And we can see that there is no public tool to provide EGFR TKI target drug annotation of gene variants automatically for nonsmall cell lung cancer samples for the moment. This is a place worth exploring in the future.
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet Tieulent J, et al. (2015) Global cancer statistics, 2012. CA: a cancer journal for clinicians 65(2): 87-108.
- Ulivi P, Chiadini E, Dazzi C, Dubini A, Costantini M, et al. (2016) Nonsquamous, Non-Small-Cell Lung Cancer Patients Who Carry a Double Mutation of EGFR, EML4-ALK or KRAS: Frequency, ClinicalPathological Characteristics, and Response to Therapy. Clin Lung Cancer 17(5): 384-390.
- Gazdar AF (2009) Activating and resistance mutations of EGFR in nonsmall-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 28 Suppl 1: S24-31.
- Westover D, Zugazagoitia J, Cho BC, Lovly CM, Paz Ares L (2018) Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol 29(suppl 1): i10-i19.
- Rossi A, Di Maio M (2015) LUX-Lung: determining the best EGFR inhibitor in NSCLC? Lancet Oncol 16(2): 118-119.
- Sutiman N, Tan SW, Tan EH, Lim WT, Kanesvaran R, et al. (2017) EGFR Mutation Subtypes Influence Survival Outcomes following First Line Gefitinib Therapy in Advanced Asian NSCLC Patients. J Thorac Oncol 12(3): 529-538.
- Suda K, Bunn PA, Jr Rivard CJ, Mitsudomi T, Hirsch FR (2017) Primary Double-Strike Therapy for Cancers to Overcome EGFR Kinase Inhibitor Resistance: Proposal from the Bench. J Thorac Oncol 12(1): 27-35.
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14): 1754-1760.
- Jo H, Koh G (2015) Faster single-end alignment generation utilizing multi-thread for BWA. Biomed Mater Eng 26 Suppl 1: S1791-S1796.
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9): 1297-1303.
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, et al. (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22(3): 568-576.
- Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16): e164.
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6(2): 80-92.
- Good BM, Ainscough BJ, McMichael JF, Su AI, Griffith OL (2014) Organizing knowledge to enable personalization of medicine in cancer. Genome Biol 15(8): 438.
- Kusnoor SV, Koonce TY, Levy MA, Lovly CM, Naylor HM, et al. (2016) My Cancer Genome: Evaluating an Educational Model to Introduce Patients and Caregivers to Precision Medicine Information. AMIA Jt Summits Transl Sci Proc 2016: 112-121.
- Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, et al. (2017) OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol.
- KW, AY, NI, J WR, TT (2017) ClinicalTrials.gov: Further Enhancements to Functionality. NLM Tech Bull (419): e9.
- Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, et al. (2017) CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet 49(2): 170-174.
- Huang L, Fernandes H, Zia H, Tavassoli P, Rennert H, et al. (2017) The cancer precision medicine knowledge base for structured clinical-grade mutations and interpretations. J Am Med Inform Assoc 24(3): 513-519.

 Mini Review
Mini Review