Abstract
FXYD3 is a Na/K-ATPase regulator which has been recently associated with different cancers development and progression; consequently, FXYD3 has been proposed in those as a potential therapeutic target. By contrast, no data are available concerning FXYD3 expression in hematological malignancies. In this study we aimed to assess FXYD3 gene expression in a large panel of B-cell derived lymphoid malignancies and to evaluate possible clinic-pathological correlations. Normal B-cell subsets served as control. We found that FXYD3 gene was not significantly modulated in normal B-cells. By contrast, BL, DLBCL, PMBCL, and PCM presented with a significant over-expression of the gene when compared to their cellular counterpart (p<0.0006). Interestingly, tumors characterized by higher FXYD3 expression presented with a significant enrichment in specific cellular functions and pathways, including NFkB pathway, WNT/B-catenin signaling, and MYC network, while FXYD3 levels also turned out to be directly related to PRDM1/BLIMP1. Finally, higher FXYD3 expression was not significantly associated with patients’ survival in PCM and DLBCL, though a trend in favour of patients with lower expression was recorded in DLBCL cases. In conclusion, we unveiled FXYD3 gene over-expression in specific non-Hodgkin lymphoma subtypes and PCM, providing evidences of its involvement in their pathobiology. Future studies are needed to define its precise role.
Keywords: FXYD3; B-cell Lymphoma; Plasma Cell Myeloma; Survival; Gene Expression; Drug Resistance
Introduction
The FXYD domain containing ion transport regulator 3 (FXYD3) belongs to the FXYD protein family, originally identified by Sweadner and Rael [1] basing on sequence similarity, and accounting in mammal for at least seven members. All these proteins have a signature sequence of six highly conserved aminoacids, comprising the FXY motif in the NH2-terminus and two glicines and one serine in the transmembrane domain. Despite that, the NH2 and COOH-termini remain variable among different members of FXYD proteins [2].
The major role of FXYD proteins consist in their ability to interact with Na/K-ATPase, adapting its activity to changing physiological demands [2]. Despite that, FXYD proteins seem to be involved in a higher number of physiological process, most of which have not been completely elucidated. One of the most interesting proteins of the FXYD family, that has been connected to tumor development and progression, is mammary tumor marker 8 (MAT-8, or FXYD3). FXYD3 was first identified in murine breast tumors initiated by NEU or RAS, and functional characterization showed that FXYD3 is capable of inducing ion-specific conductance when overexpressed in Xenopus oocytes.
While in normal tissue FXYD3 is mainly expressed in the colon, stomach, uterus and skin, it was also found to be expressed in breast tumor, in prostate tumor and in colorectal tumor. Noteworthy, it has been shown that siRNA-mediated inhibition of FXYD3 expression causes a reduction of cell proliferation in prostate cancer cell lines, so FXYD3 is probably also directly or indirectly involved in cell proliferation and it does not simply modulate the activity of Na/K-ATPase.
Different researchers are now attempting to determine the importance of FXYD3 in numerous type of cancers. In this regard, Loftas and Colleagues demonstrated that strong expression of the membrane protein FXYD3 was associated with infiltrative tumor growth and a reduction in tumor necrosis in rectal cancer, while tumors with weak FXYD3 expression had better prognosis after radiotherapy [3]. Another study proposed that a strong FXYD3 expression might help in protecting the Na/K-ATPase from the high level of oxidative stress characteristic of many tumors and also induced by cancer treatment in breast cancer cell lines [4]. It is also interesting to notice that, regarding breast cancer, the administration of estrogen and tamoxifen has been connected with the up-regulation of FXYD3 [5]. Very recently, FXYD3 has been also proposed as potential target for innovative therapeutic approaches in prostate cancer. In this study, we aimed to determine the expression and potential role of FXYD3 in B-cell derived human lymphomas.
Materials and Methods
Case Series
We studied the expression of FXYD3 gene in a discovery cohort of 277 cases for which we previously generated Gene Expression Profiles (GEP) (GSE12195 [6]; GSE4732 [7]; GSE16455 [8]; GSE12453 [9]; GSE35426 [10]; GSE24080 [11-13]. Specifically, we analyzed 46 Burkitt Lymphoma (BL), 46 Diffuse Large B-Cell Lymphoma (DLBCL), 40 Follicular Lymphoma (FL), 24 Marginal Zone Lymphoma (MZL), 22 Mantle Cell Lymphoma (MCL), 27 Chronic Lymphocytic Leukemia (CLL), 12 Classical Hodgkin Lymphoma (cHL), 5 nodular lymphocyte predominant HL, and 10 Plasma Cell Myeloma (PCM) cases; furthermore, 20 samples representative of normal B-cell subsets (10 germinal center, GC, 5 naïve, 5 Memory, and 5 plasma cells samples) were included [14].
Gene Expression Analyses
Gene expression analysis was carried out as previously reported concerning supervised, unsupervised and gene set enrichment analyses [12,13,15-19]. Briefly, the expression value of each selected gene was normalized to have a zero mean value and unit standard deviation. The distance between two individual samples was calculated by Pearson correlation with the normalized expression values. Unsupervised clustering was generated using a hierarchical algorithm based on the average-linkage method. To perform the supervised gene expression analysis, we used Gene Spring GX 12 (Agilent Technologies, Santa Clara, CA, USA). Differentially expressed genes between different groups were identified using a two-tails Student t-test and adjusted BenjaminiHochberg correction for false discovery rate, applying the following filtering criteria: p-value <0.05, and fold change>2.
Gene Set Enrichment Analysis (GSEA) of the interested gene sets was performed in the terms of Gene Ontology (GO) Biological Processes, Curated gene sets and Hallmark genes using GSEA MsigDB (www.broadinstitute.org/gsea/msigdb) web-based analysis tool [19], setting the options to the default (displaying top 10 gene sets with FDR q-value below 0.05). All data were obtained using Affymetrix HG-U133 2.0 plus microarrays (Affymetrix, Inc. http://www.affymetrix.com/support/index.affx. When we focused our analysis on FXYD3 expression, we identified FXYD3 expression using two different probe sets (202488_s_at; and 20489_s_at) in the HG-U133 2.0 plus microarray. The median value from the two probes was used for the analysis.
Statistical Analysis
Statistical analyses were performed using IBM SPSS Statistics 20.0 and Prism (GraphPad softwares, USA). ANOVA and unpaired T-tests were used for continuous variables examination. When a sample size was less than 10 cases in at least 1 group a nonparametric (Mann-Whitney) test was used to analyze the GEP data to compare FXYD3 expression in different subgroups. Survival analyses were performed by Kaplan-Meier method. Two-sided tests were used in all calculations. The limit of significance for all analyses was defined as p<0.05. Survival analyses for DLBCL and PCM patients were carried out using the GSE34171 and GSE24080 datasets, respectively.
Results
FXYD3 is Over-Expressed in Burkitt lymphoma, Diffuse Large B-Cell Lymphoma, Plasma Cell Myeloma And Primary Mediastinal B-Cell Lymphoma
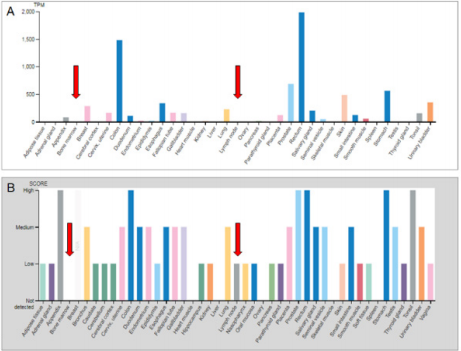
First of all, we investigated FXYD3 expression in normal tissues including bone marrow and lymph nodes by referring to the Human Protein Atlas (https://www.proteinatlas.org/ENSG00000089356- FXYD3/tissue). We observed that non neoplastic hematopoietic tissues presented with a very limited expression at both mRNA and protein level. Conversely, gastro-intestinal tract as well as prostate presented the highest expression scores (Figure 1A & 1B).
Figure 1: FXYD3 gene expression in lymphoma subtypes at mRNA (A) and protein (B) levels. Data obtained from The Human Protein Atlas.
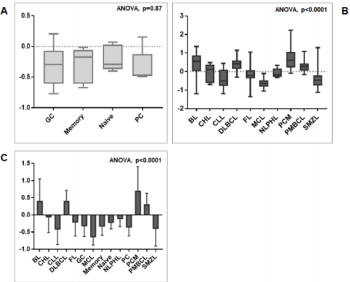
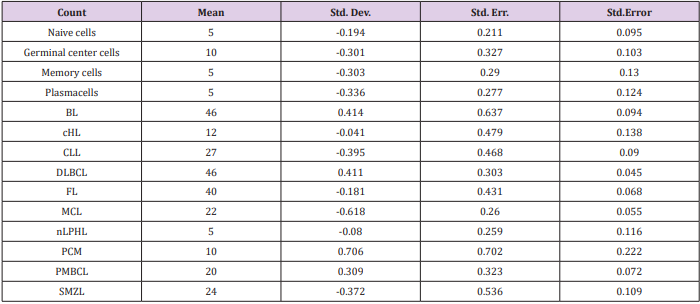
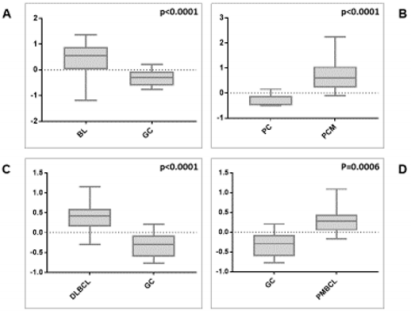
We then sought to analyse the expression of FXYD3 gene in different phases of B-cells development, including germinal centre (GC), memory B cell, naïve B cell and plasma cells, without finding any significant modulation throughout the germinal centre transition (ANOVA, p=0.87; Figure 2A). Following, we studied the expression of FXYD3 gene in a large panel of lymphoid malignancies, including Hodgkin and non-Hodgkin lymphomas, CLL and PCM. The variance analysis showed a quite significant variability among samples (ANOVA, p<0.0001; Figure 2B). Particularly, the germinal center derived aggressive lymphomas (BL, DLBCL, and PMBCL), as well as PCM showed the highest expression values across the panel (Figure 2C). Indeed, the direct comparison of tumors vs. their corresponding cellular counterpart, indicated a significant overexpression of FXYD3 gene (Figure 3A-3D). Detailed expression values of FXYD3 gene in the different categories are reported in Table 1.
Figure 2: FXYD3 gene expression in normal and neoplastic samples. A) FXYD3 gene expression in non-neoplastic B-cell subsets; B) FXYD3 gene expression in lymphomas, CLL and PCM; C) Interaction bar plot indicative of subsets with the highest expression values.
FXYD3 Over-Expression is Associated with Specific Pathways and Cellular Programs
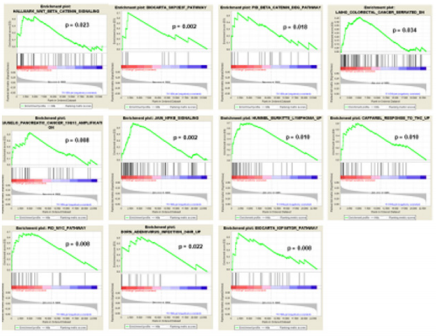
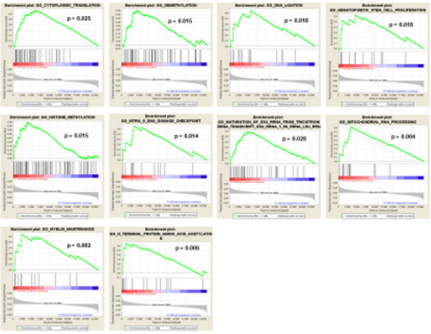
In order to determine the possible role that the overexpression of FXYD3 gene has for BL, DLBCL, PCM and PMBCL, we investigated whether FXYD3 over-expression was paralleled by over-expression of genes involved in specific cellular functions and/or pathways. To address this issue, we first compared by Gene Set Enrichment Analysis (GSEA) cases that, irrespectively of their histology, presented with the highest (above the 75th percentile) and the lowest (below the 25th percentile) FXYD3 gene expression values. We identified 112 gene sets differentially expressed in the two groups (p<0.05) (Table 2). Among others, we observed that cases presenting with the highest FXYD3 expression presented with significant enrichment of genes related to WNT/B-catenin signaling, NFKB signaling, MYC pathway, IGF1/MTOR pathway and adenovirus infection (Figure 3). Further, the highest FXYD3 expression was associated with epigenetic reprogramming, hematopoietic stem cell proliferation, DNA damage check point and protein acetylation (Figure 4).
Figure 3: FXYD3 gene over-expression in BL, DLBCL, PMBCL and PCM, in contrast with their normal counterparts (panels A, B, C, and D, respectively).
Table 2: Gene set enrichment analysis (GSEA) of cases with higher or lower (75° vs 25°) FXYD3 gene expression (top 20 are shown).
FXYD3 Expression Depends on PRDM1/BLIMP1
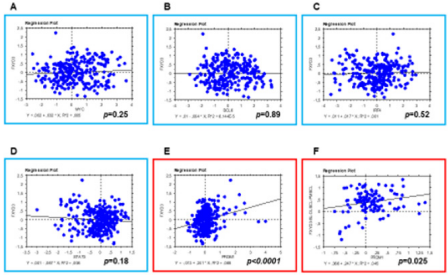
In order to assess whether FXYD3 gene expression was related to any specific transcription factor known to be involved in B-cell tumorigenesis, we studied the expression of BCL6, MYC, IRF4, PRDM1/BLIMP1, and STAT3 and correlated it to FXYD3. We found that PRDM1/BLIMP1 expression was significantly correlated with FXYD3one (p<0.0001); by contrast, BCL6, MYC, IRF4, and STAT3did not present any significant degree of correlation with FXYD3 (Figures 5 & 6). The correlation was still significant when only BL, DLBCL, PMBCL, and PCM (i.e. the tumors in which FXYD3 over-expression was documented) were considered. However, since PRDM1/ BLIMP1 is consistently expressed in PCM, as in non-neoplastic plasma cells, we then intended to analyze BL, DLBCL, and PMBCL only. Again, the correlation was highly significant (p=0.025; Figure 6F).
Figure 5: GSEA – FXYD3 High vs. Low (75° vs 25°) – Selected Gene Ontology Biological Processes gene sets are plotted.
Figure 6: FXYD3 expression is related to PRDM1/BLIMP1. Correlation between FXYD3 and MYC (A), BCL6 (B), IRF4 (C), and STAT3 (D) expression; all samples included. Correlation between FXYD3 and PRDM1: E) All samples included; F) Only BL, DLBCL, and PMBCL included.
FXYD3 Over-Expression is not Related to Clinical Aggressiveness And Survival in DLBCL and PCM
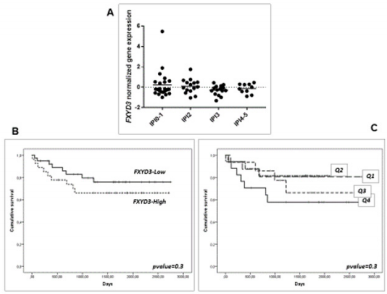
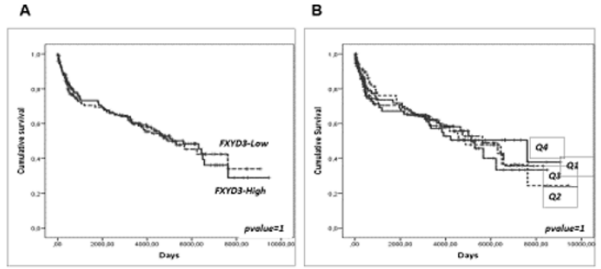
Last, we sought to determine whether the over-expression of FXYD3 gene could be related to clinical aggressiveness and survival in DLBCL and PCM, large panel of cases provided with clinical information being available for these two diseases but not for PMBCL and BL. While some studies have shown that the expression of FXYD3 may be capable of influencing clinical aggressiveness and prognosis in other tumors [3], we failed to identify any significant correlation. Particularly, in DLBCL, the International Prognostic Index (IPI), the most widely used clinical parameter to predict DLBCL outcome, was not significantly associated with FXYD3 gene expression (Figure 6A). Consistently, it was not significantly associated with the Overall Survival (OS), though a slight trend in favor of patients with the lowest values could be observed (Figure 6B-C). We cannot exclude, however, that in larger data sets provided with complete molecular (i.e. ABC vs. GCB), genetic (MYC/ BCL2/BCL6 abnormalities) and phenotypic (double expressor vs. standard) it might be possible to detect significant correlations. In PCM patients, FXYD3 gene expression was not related to the OS, nor any trend could be noted (Figure 7A-B).
Figure 7: Prognostic value of FXYD3 gene expression in DLBCL. A) association with the IPI; B) association with the overall survival (above vs. below the 50° percentile); C) association with the overall survival by quartiles of expression.
Discussion
Our study aimed to determine for the first time the expression of FXYD3gene in non-neoplastic B-cell subsets as well as in different types of B-cell neoplasms. We observed a significant over-expression of the gene in aggressive germinal center derived lymphomas (namely, BL, DLBCL, and PMBCL), as well as in PCM. This expression pattern was definitely interesting. In fact, on the one hand it did not directly reflect tumor aggressiveness, since MCL showed pretty low values while PCM showed rather high ones. On the other hand, it did not reflect the cellular origin, since neither FL nor HL, two germinal center derived neoplasms, did parallel the expression patterns observed in the other GC-derived malignancies. Therefore, although FXYD3 expression tended to be higher in highly proliferating lymphomas (BL, DLBCL, and PMBCL), its over-expression, documented also in PCM, a tumor generally characterized by a lower proliferating rate, did not appear to be related to a single feature but rather to be referred to specific single entities (Figure 8).
Figure 8: Prognostic value of FXYD3 gene expression in DLBCL. A) association with the IPI; B) association with the overall survival (above vs. below the 50° percentile); C) association with the overall survival by quartiles of expression.
We then sought to understand whether FXYD3 over-expression was associated with the activation of specific cellular functions. To do this, we analyzed by GSEA the gene signature associated with FXYD3 over-expression and found several pathways relevant for malignant transformation being involved. Of interest, the enrichment in genes related to WNT/ -catenin signaling is similar to what observed in colorectal cancer [20], where the pathway of APC and -catenin is a well-known feature, of fundamental importance in the development of this tumor type. In addition, it is noteworthy that the expression of FXYD3 is also connected to modifications of physiological mechanisms of epigenetic regulation. In fact, demethylating agents as well as histone deacetylase inhibitors are currently in clinical use in haematological malignancies [21] and their application/efficacy might be enhanced in tumors carrying FXYD3 over-expression. Therefore, it might be useful to evaluate FXYD3 gene expression in patients who received epigenetic modifiers and correlate it to the clinical response and, in case of positive evidences, to test FXYD3 as potential biomarkers prospectively.
Grippingly, FXYD3 expression was found to be directly related to that of PRDM1 [22]. This gene encodes for BLIMP1 protein, a transcription factor necessary for plasma cell differentiation [23]. In fact, while PAX5 is critical for the maintenance of B cell identity [24], the expression of BLIMP1 leads to the repression of both PAX5 and BCL6and it is capable of triggering plasma cell differentiation by ensuring that plasma cell cannot return to an earlier developmental stage [25]; BLIMP1, also, has been correlated with the induction of genes that are involved in the immunoglobulin secretion [26]. Indeed, our correlation between FXYD3 and PRDM1 is consistent with the evidence that FXYD3 expression was not significantly associated with the clinical outcome in terms of overall survival in PCM. In fact, PRDM1 is always expressed in this tumor, reflecting its physiological role in plasma cells. By contrast, PRDM1 expression was correlated with poor prognosis in ABC-type DLBCL [27]; however, we failed to document significant differences among DLBCL cases based on FXYD3 expression.
The main limitation of this study probably relies on the fact that all analyses were conducted by GEP only and were not supported by an additional method, due to the lack of available pathological material. In this regard, a couple of consideration must be taken into account. Firstly, GEP, at least when performed by Affymetrix microarrays, is now considered to be a robust and reliable method to measure and quantify genes expression in a given sample, with substantial correspondence (despite an inferior sensitivity) to quantitative PCR methods [28]. Therefore, a validation at protein level would be the most appropriate. However, a number of studies previously sought to compare GEP and IHC (the unique proteomic technique feasible in formalin fixed paraffin embedded tissue samples) [29,30] demonstrating a high degree of consistency between the two methods (around 77-87%) that made such validation approaches even unnecessary in many applications [31,32]. Moreover, IHC might even fail in detecting subtle but still biologically significant differences caught at gene expression level [33]. It would be interesting, instead, to evaluate in future independent series whether GEP or IHC will be able to stratify lymphoma and/or myeloma patients with different clinicopathological features based on FXYD3 expression [34].
Conclusion
In conclusion, we unveiled the over-expression of FXYD3 gene in some diseases (BL, DLBCL, PCM, and PMBCL), and connected its expression to the activation of specific cellular pathways and a well-known transcription factor. On the clinical ground, FXYD3 over-expression was not significantly correlated to overall survival in DLBCL and PCM, though different attempts might be warranted. Finally, based on the molecular profile associated to FXYD3 gene over-expression, the specific association of this molecule with clinical response to epigenetic modifiers might be tested.
Acknowledgements
This work was supported by Istituto Euro-Mediterraneo di Scienza e Tecnologia (IEMEST), AIL Pesaro ONLUS, RFO (Prof. Piccaluga), FIRB Futura 2011 RBFR12D1CB (Prof. Piccaluga), and Fondi Dipartimentali RFO 2014-2018 (Prof. Piccaluga).
References
- Sweadner KJ, Rael E (2000) The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics 68(1): 41-56.
- Crambert G, Geering K (2003) FXYD proteins: new tissue-specific regulators of the ubiquitous Na, K-ATPase. Sci STKE 2003(166): RE1.
- Loftas P, Arbman G, Sun XF, Edler D, Syk E, et al. (2016) FXYD-3 expression in relation to local recurrence of rectal cancer. Radiat Oncol J 34(1): 52-58.
- Liu CC, Teh R, Mozar CA, Baxter RC, Rasmussen HH (2016) Silencing overexpression of FXYD3 protein in breast cancer cells amplifies effects of doxorubicin and gamma-radiation on Na(+)/K(+)-ATPase and cell survival. Breast Cancer Res Treat 155(2): 203-213.
- Herrmann P, Aronica SM (2015) Estrogen and tamoxifen up-regulate FXYD3 on breast cancer cells: assessing the differential roles of ER alpha and ZEB1. Springerplus 4: 245.
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, et al. (2009) Mutations of multiple genes cause deregulation of NF-kappa B in diffuse large B-cell lymphoma. Nature 459(7247): 717-721.
- Dave SS, Fu K, Wright GW, Lam LT, Kluin P, et al. (2006) Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med 354(23): 2431-2442.
- Fernandez V, Salamero O, Espinet B, Sole F, Royo C, et al. (2010) Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res 70(4): 1408-1418.
- Brune V, Tiacci E, Pfeil I, Doring C, Eckerle S, et al. (2008) Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med 205(10): 2251-2268.
- Watkins AJ, Hamoudi RA, Zeng N, Yan Q, Huang Y, et al. (2012) An integrated genomic and expression analysis of 7q deletion in splenic marginal zone lymphoma. PLoS One 7(9): 44997.
- Shi L, Campbell G, Jones WD, Campagne F, Wen Z, et al. (2010) The Micro Array Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol 28(8): 827-838.
- Piccaluga PP, Califano A, Klein U, Agostinelli C, Bellosillo B, et al. (2008) Gene expression analysis provides a potential rationale for revising the histological grading of follicular lymphomas. Haematologica 93(7): 1033-1038.
- Piccaluga PP, De Falco G, Kustagi M, Gazzola A, Agostinelli C, et al. (2011) Gene expression analysis uncovers similarity and differences among Burkitt lymphoma subtypes. Blood 117(13): 3596-3608.
- Piccaluga PP, Agostinelli C, Fuligni F, Righi S, Tripodo C, et al. (2015) IFI16 Expression Is Related to Selected Transcription Factors during B-Cell Differentiation. J Immunol Res 2015: 747645.
- Piccaluga PP, Agostinelli C, Califano A, Rossi M, Basso K, et al. (2007) Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest 117(3): 823-834.
- Piccaluga PP, Agostinelli C, Califano A, Carbone A, Fantoni L, et al. (2007) Gene expression analysis of angioimmunoblastic lymphoma indicates derivation from T follicular helper cells and vascular endothelial growth factor deregulation. Cancer Res 67(22): 10703-10710.
- Piccaluga PP, Fuligni F, De Leo A, Bertuzzi C, Rossi M, et al. (2013) Molecular profiling improves classification and prognostication of nodal peripheral T-cell lymphomas: results of a phase III diagnostic accuracy study. J Clin Oncol 31(24): 3019-3025.
- Navari M, Fuligni F, Laginestra MA, Etebari M, Ambrosio MR, et al. (2014) Molecular signature of Epstein Barr virus-positive Burkitt lymphoma and post-transplant lymphoproliferative disorder suggest different roles for Epstein Barr virus. Front Microbiol 5: 728.
- Navari M, Etebari M, De Falco G, Ambrosio MR, Gibellini D, et al. (2015) The presence of Epstein-Barr virus significantly impacts the transcriptional profile in immunodeficiency-associated Burkitt lymphoma. Front Microbiol 6: 556.
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, et al. (1997) Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275(5307): 1787-1790.
- Markozashvili D, Ribrag V, Vassetzky YS (2015) Histone deacetylase inhibitors and epigenetic regulation in lymphoid malignancies. Invest New Drugs 33(6): 1280-1291.
- Boi M, Zucca E, Inghirami G, Bertoni F (2015) PRDM1/BLIMP1: a tumor suppressor gene in B and T cell lymphomas. Leuk Lymphoma 56(5): 1223-1228.
- Fairfax KA, Kallies A, Nutt SL, Tarlinton DM (2008) Plasma cell development: from B-cell subsets to long-term survival niches. Semin Immunol 20(1): 49-58.
- Schebesta M, Heavey B, Busslinger M (2002) Transcriptional control of B-cell development. Curr Opin Immunol 14(2): 216-223.
- Shapiro-Shelef M, Calame K (2005) Regulation of plasma-cell development. Nat Rev Immunol 5(3): 230-242.
- Shaffer AL, Shapiro Shelef M, Iwakoshi NN, Lee AH, Qian SB, et al. (2004) XBP1, Downstream of Blimp-1, Expands the Secretory Apparatus and Other Organelles, and Increases Protein Synthesis in Plasma Cell Differentiation. Immunity 21(1): 81-93.
- Xia Y, Xu Monette ZY, Tzankov A, Li X, Manyam GC, et al. (2017) Loss of PRDM1/BLIMP-1 function contributes to poor prognosis of activated B-cell-like diffuse large B-cell lymphoma. Leukemia 31(3): 625-636.
- Maruyama K, Yamaguchi Shinozaki K, Shinozaki K (2014) Gene expression profiling using DNA microarrays. Methods Mol Biol 1062: 381-391.
- Gutierrez Garcia G, Cardesa Salzmann T, Climent F, Gonzalez Barca E, Mercadal S, et al. (2011) Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood 117(18): 4836-4843.
- Hwang HS, Park CS, Yoon DH, Suh C, Huh J (2014) High concordance of gene expression profiling-correlated immunohistochemistry algorithms in diffuse large B-cell lymphoma, not otherwise specified. Am J Surg Pathol 38(8): 1046-1057.
- Meyer PN, Fu K, Greiner TC, Smith LM, Delabie J, et al. (2011) Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol 29(2): 200-207.
- Lin F, Chen Z (2014) Standardization of diagnostic immunohistochemistry: literature review and geisinger experience. Arch Pathol Lab Med 138(12): 1564-1577.
- Scott DW, Wright GW, Williams PM, Lih CJ, Walsh W, et al. (2014) Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood 123(8): 1214-1217.
- Altman N, Krzywinski M (2016) P values and the search for significance. Nature Methods 14: 3-4.

 Research Article
Research Article