Abstract
Purpose: The aim of this study was to find the prevalence of Candida species isolated from vaginal discharge of women undergoing IVF-ET in Vietnam.
Materials and Methods: 83 strains isolated from women undergoing IVF-ET in Military Institute of Clinical Embryology and Histology were collected between November 2017 and December 2018. 83/83 strains were identified as Candida species by several phenotypic methods (i.e., germ tube test, culture on Brilliance™ Candida Agar) and molecular (PCR-RFLP and sequencing).
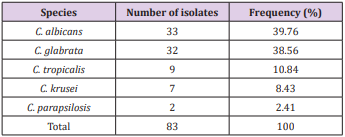
Results: The results showed that, C. albicans (n = 33, 39.76%) was the most common species, followed by C. glabrata (n = 32, 38.56%), C. tropicalis (n = 9, 10.84%), C. krusei (n = 7, 8.43%), C. parapsilosis (n = 2, 2.41%), respectively.
Conclusion: In the present study, the most common species in women undergoing IVF-ET were C. albicans, C. glabrata and C. tropicalis, respectively. The frequency of the non-albicans Candida species were increasing, especially C. glabrata.
Keywords: Prevalence; Candida; IVFET; Vietnam
Abbreviations: VVC: Vulvovaginal Candidiasis; NAC: Non-Albicans Candida; SDA: Sabouraud’s Dextrose Agar; RFLP: Restriction Fragment Length Polymorphism
Introduction
Vulvovaginal candidiasis is estimated to be the second most common cause of vaginitis after bacterial vaginosis [1]. Many studies have shown that approximately 75% of healthy female population at least one symptomatic episode of vulvovaginal candidiasis (VVC) and 40-50% will have recurrent episodes during their lifetime [2,3]. Candida albicans was accounted as the first vulvovaginal candidiasis causative agent [1], followed by non-albicans Candida (NAC) species including Candida glabrata, Candida parapsilosis, Candida tropicalis and Candida krusei [4]. The species distribution of Candida isolates varies between countries, regions, and institutions [5]. NAC species have increasingly been identified as the cause of vulvovaginitis [6]. The data of some reports indicated that NAC species were responsible for 10% to 30% of episodes in certain geographic regions [6]. In clinical practice, the yeast identification is based on morphological and biochemical markers, including the automated methods [7]. However, not all the species are precisely identified by such procedures. Therefore, molecular markers were utilized in the present study to enable identification and detection of various strains [8]. According to the literature, identification of the Candida isolates is important for treatment, prediction of the prognosis, performing infection control, finding epidemiological data and avoid antifungal failure [9,10]. In addition, the species identification of the yeast should be performed with a high accuracy [11]. Previously, very limited data is available on species composition of Candida in Vietnam, especially the women undergoing IVF-ET. This study has been performed to find the prevalence of Candida species isolated from vaginal discharge of women undergoing IVF-ET in Vietnam.
Materials and Methods
Yeast Strains
A total of 83 strains of Candida species isolated from women undergoing IVF-ET in Military Institute of Clinical Embryology and Histology, Vietnam Military Medical University were used. The isolates were cultured on Sabouraud’s Dextrose Agar (SDA) for 48h at 30o C and suspended in sodium chloride solution 0.85% at a concentration of McFarland 0.5 (corresponds to 1-5 x 106 cfu/ ml). In this study, C. albicans (ATCC 90028) was used as a standard quality control strain.
Yeast Identification
The yeasts were first identified according to morphological and physiological characteristics using germ tube test (in human serum), cultured on Brilliance™ Candida Agar (Oxoid, UK), and microscopic morphology on cornmeal agar (Difco, USA) with 1% Tween 80 (Merck, Germany).
DNA Extraction
Genomic DNA were extracted from yeast cultures using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany), following manufacture recommendation. DNA concentration (ng/μl) was estimated using a NanoDropTM 2000 Spectrophotometer at 260 nm (Thermo Fisher Scientific, USA).
PCR Reactions
To amplify an approximate 500-900 bp region of the ITS1- 5.8S-ITS2 sequence, PCR was performed using a set of ITS1 (5´- TCC GTA GGT GAA CCT GCG G -3´) and ITS4 (5´-TCC TCC GCT TAT TGA TAT GC-3´) (Integrated DNA Technologies, USA) as sense and antisense primers (Mirhendi et al., 2006) [12], respectively. Total volume of PCR reaction was 50μl containing 5μl of DNA solution, 25 μl mastermix 2X (Thermo Fisher Scientific, USA), 1.0μl of each primer (0.2μM) and 18.0μl of distilled water (Corning, USA). PCR amplification was performed in Thermo Mastercycler Gradient (Thermo Fisher Scientific, USA). The reaction cycle was as follows: an initial denaturation step at 94°C for 5 minutes, followed by 35 cycles of 94°C for 30 seconds, 56°C for 45 seconds and 72°C for 60 seconds and a final extension of 72°C for 15 minutes.
Restriction Fragment Length Polymorphism (RFLP) Analysis
Restriction fragment length polymorphism was performed according to the method described by Mirhendi et al. to identify the most medically important Candida species [12]. To perform RFLP assay, total volume of 16μl, including 5μl PCR product was added with 1μl of MspI (10UI), 1μl of 10X Tango buffer (Thermo Fisher Scientific, USA) and 9μl of distilled water. According to the manufacturer’s instruction, the tubes were incubated at 37°C for 3 hours, to make sure full cutting of fragments. For analyzing the digestion products, 6μl of each product in addition to 1μl of loading dye buffer were electrophoresed on 2% agarose gel in 1X TBE buffer for about 2.0h at 90V and visualized staining with 0.5µg/ml of ethidium bromide on UV illumination (UVP, Canada). The size of each band was determined by a 50bp plus ladder molecular weight marker (Thermo Fisher Scientific, USA).
D1/D2 26S rDNA Gene Sequencing
Amplification of the D1/D2 domains of the 26S rRNA gene were achieved with the primers NL-1 (5’-GCA TAT CAA TAA GCG GAG GAA AAG-3’) and NL-4 (5’-GGT CCG TGT TTC AAG ACG G-3’). PCR products of D1/D2 from three isolates (HN-SD20, HN-SD42 and HN-SD51) were sent to First BASE Laboratories Sdn Bhdservice (Kembangan 43300, Selangor, Malaysia) for purification and automatic sequencing in both directions, using the same primers which were used in the PCR. Sequences were read on ABI 3130 Genetic Analyzer software (Seq Scape 2.1). The accuracy of data was confirmed by two-directional sequencing. Representative sequences were deposited in the GenBank under accession number MK464263, MK464264 and MK464265.
Data and Sequence Analyses
IBM SPSS statistics 20.0 software was used for data processing in this study. The sequences generated were compared to available data in the NCBI database using BLAST guidelines.
Results
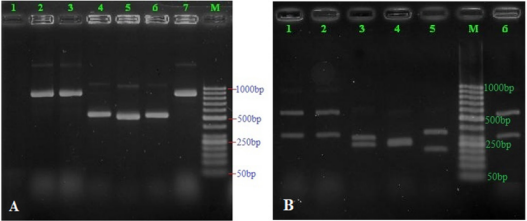
A total of 83 Candida strains were isolated during the study period. All of them came from Military Institute of Clinical Embryology and Histology, Vietnam Military Medical University. The overall species distribution is shown in Table 1. The frequency of non-Candida albicans species was 60.24%. Distribution of Candida species isolated from vaginal discharge of women undergoing IVFET was as follows: 33 C. albicans (39.76%), 32 C. glabrata (38.56%), 9 C. tropicalis (10.84%), 7 C. krusei (8.43%) and 2 C. parapsilosis (2.41%) (Table 1). C. albicans was the most commonly isolated species in our study. The second leading strain was C. glabrata. All cases were monomicrobial infections, no mixed infections were detected in the present study (Figure 1a & 1b). DNA sequencing of three D1/D2 were compared to available data in the NCBI database using BLAST guidelines. BLAST results showed that C. tropicalis from China (MG871744.1) possessed the sequence most like those in these 2 isolates with 100% identity for HN-SD20 isolate (MK464263.1) and HN-SD51 isolate (MK464265.1). D1/D2 sequencing of HNSD42 strain was 100% like C. albicans from China (KY825125.1).
Figure 1: Patterns of PCR products of Candida isolates before (Figure 1a) and after digestion with the restriction enzyme MspI (Figure 1b). Lane M: 50 bp ladder molecular weight marker; land 1 (A): negative control; land 2-7 denoted to different Candida samples amplified as a single band of 500-900 bp; land 1-2 and 6 (B) denoted to those of C. glabrata; land 3 (B) denoted to these of C. albicans; land 4 (B) denoted to these of C. krusei; land 5 (B) denoted to these of C. tropicalis.
Table 1: Frequency of different Candida species isolated from vaginal discharge of women undergoing IVF-ET.
Discussion
There are many techniques that have been proposed to detect and distinguish Candida species in clinical samples [13]. In present study, we used three phenotypic methods and two molecular methods, namely PCR-RFLP and DNA sequencing, to identify Candida species in 83 strains from vaginal discharge of women undergoing IVF-ET. The results of our study showed that C. albicans continues to be the species most commonly isolated from vaginal discharge of women. The result from our study was like that of some previous researches in China [9,14], Iran [3,15] and Brazil [8]. Although C. albicans was still the most frequently isolated species, we describe a very high proportion of non-albicans Candida species (60.24%), with C. glabrata, being almost as common as C. albicans. C. tropicalis was the third most frequently isolated Candida species (10.84%), followed by C. krusei (8.43%). In this series we found only two C. parapsilosis (2.41%). The results of our study were like that of some previous researches.
Most reports have shown that C. glabrata, C. tropicalis and C. krusei were accounted as the second, third and fourth common agents of disease, respectively [16,17]. According to the results from our study, C. glabrata was the second most commonly occurring species in Vietnam. Abbasi et al. (2017) reported similar findings in Iran, where C. glabrata was also the second most common species isolated from vulvovaginal Candidiasis [15]. Conversely, a report from India indicated that C. glabrata was the most common species which was isolated from 50.4% patients with vulvovaginal candidiasis [16]. The reasons for such differences in the distribution of Candida spp. are not fully understood. However, the high prevalence of C. albicans and C. glabrata in our study was remarkable results. According to Castrillón-Duque (2018), the presence of C. albicans or C. glabrata affects seminal parameters [18]. The data from this study showed that more studies are needed to determine the effects of vaginal yeast infections on results of IVF-ET.
Conclusion
The most common species in women undergoing IVF-ET were C. albicans, C. glabrata and C. tropicalis, respectively. The frequency of the non-albicans Candida species isolated from women undergoing IVF-ET is gradually increasing, especially C. glabrata.
Acknowledgement
This study was supported by Military Medical University, Hanoi, Vietnam. The authors declare that there is no conflict of interest.
References
- Martin Lopez JE (2015) Candidiasis (vulvovaginal). BMJ clinical evidence 0815.
- Mendling W, Brasch J, Cornely OA, Effendy I, Friese K, et al. (2015) Guideline: vulvovaginal candidosis (AWMF 015/072), S2k (excluding chronic mucocutaneous candidosis). Mycoses 58(1): 1-15.
- Hedayati MT, Taheri Z, Galinimoghadam T, Aghili SR, Yazdani Cherati J, et al. (2015) Isolation of different species of Candida in patients with vulvovaginal candidiasis from Sari, Iran. Jundishapur journal of microbiology 8(4): e15992-e15992.
- Ilkit M, Guzel AB (2011) The epidemiology, pathogenesis, and diagnosis of vulvovaginal candidosis: a mycological perspective. Critical reviews in microbiology 37(3): 250-261.
- Samra Z, Yardeni M, Peled N, Bishara J (2005) Species distribution and antifungal susceptibility of Candida bloodstream isolates in a tertiary medical center in Israel. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology 24(9): 592-595.
- Kennedy MA, Sobel JD (2010) Vulvovaginal Candidiasis Caused by Nonalbicans Candida Species: New Insights. Current infectious disease reports 12(6): 465-470.
- Lockhart SR, Messer SA, Pfaller MA, Diekema DJ (2008) Geographic distribution and antifungal susceptibility of the newly described species Candida orthopsilosis and Candida metapsilosis in comparison to the closely related species Candida parapsilosis. Journal of Clinical Microbiology 46(8): 2659-2664.
- Fornari G, Vicente VA, Gomes RR, Muro MD, Pinheiro RL, et al. (2016) Susceptibility and molecular characterization of Candida species from patients with vulvovaginitis. Brazilian journal of microbiology: [publication of the Brazilian Society for Microbiology] 47(2): 373-380.
- Shi XY, Yang YP, Zhang Y, Li W, Wang JD, et al. (2015) Molecular identification and antifungal susceptibility of 186 Candida isolates from vulvovaginal candidiasis in southern China. Journal of medical microbiology 64(Pt 4): 390-393.
- Korkmaz P, Alpay Y, Çevik F, Aykin N, Koygun A, et al. (2015) Distribution and antifungal resistance of Candida species isolated from intensive care units. Turkish Journal of Medical Sciences 45: 740-741.
- Sadrossadati SZ, Ghahri M, Imani Fooladi AA, Sayyahfar S, Beyraghi S, et al. (2018) Phenotypic and genotypic characterization of Candida species isolated from candideamia in Iran. Current medical mycology 4(2): 14- 20.
- Mirhendi H, Makimura K, Khoramizadeh M, Yamaguchi H (2006) A oneenzyme PCR-RFLP assay for identification of six medically important Candida species. Japanese journal of medical mycology 47(3): 225-229.
- Madhavan PFJ, Pei PC (2011) Laboratory Isolation and identification of Candida species. Journal of applied Sciences 11(16): 2870-2877.
- Wang FJ, Zhang D, Liu ZH, Wu WX, Bai HH, et al. (2016) Species Distribution and In Vitro Antifungal Susceptibility of Vulvovaginal Candida Isolates in China. Chinese medical journal 129(10): 1161-1165.
- Abbasi Nejat Z, Farahyar S, Falahati M, Ashrafi Khozani M, Hosseini et al. (2017) Molecular Identification and Antifungal Susceptibility Pattern of Non-albicans Candida Species Isolated from Vulvovaginal Candidiasis. Iranian biomedical journal 22(1): 33-41.
- Mohanty S, Xess I, Hasan F, Kapil A, Mittal S, et al. (2007) Prevalence & susceptibility to fluconazole of Candida species causing vulvovaginitis. The Indian journal of medical research 126(3): 216-219.
- Rezaei Matehkolaei A, Shafiei S, Zarei Mahmoudabadi A (2016) Isolation, molecular identification, and antifungal susceptibility profiles of vaginal isolates of Candida species. Iranian journal of microbiology 8(6): 410- 417.
- Castrillon Duque EX, Puerta Suarez J, Cardona Maya WD (2018) Yeast and Fertility: Effects of In Vitro Activity of Candida spp. on Sperm Quality. Journal of reproduction & infertility 19(1): 49-55.

 Research Article
Research Article