Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Masato Kantake*
Received: August 16, 2018; Published: August 27, 2018
*Corresponding author: Masato Kantake, Neonatal Medical Center, Juntendo University Shizuoka Hospital, 1192 Nagaoka, Izunokuni, Shizuoka, 410-2295, Japan
DOI: 10.26717/BJSTR.2018.08.001648
Fetal nucleated cells within maternal blood represent a potential source of fetal genes obtainable by venipuncture. In our previous study, we have enriched CD45-CD71+ cells from maternal blood. To confirm the potential usefulness of our method, we analyzed the origin of the enriched CD45-CD71+ cells using a real-time polymerase chain reaction (PCR) system.
The SRY (sex-determining region Y) gene was detected only in cells extracted from the blood of the pregnant woman with the male fetus. The relative dose of the SRY gene to the TFRC (transferrin receptor) gene was 0.001, indicating that 99.9% of the separated cells were from the mother and only 0.1% were of fetal origin.
Our method using MACS technology was insufficient to analyze the fetal genome, methylome, or transcriptome, as some additional enrichment is needed, such as single cell technology
Fetal nucleated cells within maternal blood represent a potential source of fetal genes obtainable by venipuncture. An analysis of nuclear red blood cells (NRBCs) in pregnant woman is believed to be useful for understanding prenatal fetal status. However, the extreme scarcity of fetal cells puts strong demands on sensitivity and specificity. We have effectively used a simple MACS technology without density gradient centrifugation or lysis of RBCs [1].
Figure 1: Flowcytometry of enriched CD45-CD71+ cells of patient 1. DNA was extracted from these CD45-CD71+ cells.
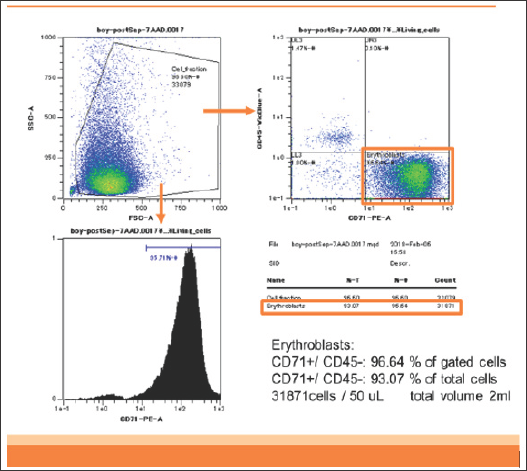
Briefly, in our previous study, we analyzed two patients. Patient 1 was an 18-year-old woman bearing a single male fetus. Patient 2 was a 42-year-old woman bearing a single female fetus. These two blood samples (2 mL each) containing anticoagulant were stored at room temperature and analyzed within 12 hours after sampling. In patient 1, CD45-CD71+ cells were enriched from 0.3% in the analyzed cells to 93% in the enriched cells (310-fold). The yield of CD45-CD71+ cells was 2.5 x 106 cells from 2 mL of maternal blood (Figure 1).
In patient 2, CD45-CD71+ cells were also enriched from 0.3% to 93%. The yield was 3.7 x 106 cells from 2 mL of maternal blood.
To confirm the potential usefulness of our method, we analyzed the origin of the enriched CD45-CD71+ cells using a real-time polymerase chain reaction (PCR) system.
a) Genomic DNA was extracted from frozen enriched CD45- CD71+ cells using the DNeasy® Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The extracted DNA (20 ng) was used in the following reaction.
b) We used three pairs of primers for the real-time PCR:
TFRC-F; 5'-ACGGAATATGAAGATCTCAGCAAGG-3'
TFRC-R; 5'-GCGCAGATCACTAACGAGATGGA-3'
HPRT1-F; 5'-GTTGGCTGAAATAGTTGAACAGCTT-3'
HPRT1-R; 5'-TGGGACTTCTGCCTCCAGAGTA-3'
HA086596 (SRY)-F; 5'-GCGAAGATGCTGCCGAAGA-3'
HA086596 (SRY)-R; 5'-TTGAGTGTGTGGCTTTC GTACAGTC-3'
TFRC: transferrin receptor gene; one copy on one autosome
HPRT1: hypoxanthine phosphoribosyl transferase gene; one copy on the X chromosome
SRY: sex-determining region Y; one copy on the Y chromosome
c) Real-time PCR was performed using the Thermal Cycler Dice® Real Time System (Takara Bio, Shiga, Japan) and SYBR®Premix Ex TaqTM reagents according to the manufacturer's protocol.
d) The relative dose of the SRY and HPRT1 genes to the TFRC gene were calculated.
Male and female are control subjects.
The values are standardized by TFRC gene dose.
The SRY gene was detected only in cells extracted from the blood of the pregnant woman with the male fetus. The relative dose of the SRY gene to the TFRC gene was 0.001, indicating that 99.9% of the separated cells were from the mother and only 0.1% were of fetal origin (Table1).
Our method using MACS technology was insufficient to analyze the fetal genome, methylome, or transcriptome, as some additional enrichment is needed, such as single cell technology [2]. Erythroblasts express the CD71 antigen on their cell surfaces from the burst-forming unit and erythroid stages up to and including the nuclear extrusion (reticulocyte) stage [3]. CD71+ cells in pregnant women contain fetal erythroblasts, a small number of maternal lymphocytes, and a particular number of reticulocytes of unknown origin. Our strategy using CD45 negative selection to avoid contamination of maternal monocytes or lymphocytes prior to CD71 positive selection was thought to be reasonable, but the reason why we obtained a relatively high yield of CD45-CD71+ cells in the previous study was because of contamination of recently enucleated maternal erythrocytes (reticulocytes). Cell-free fetal (cff) DNA provided some information for a genome or methylome analysis. It is known that a relatively high dose of cff DNA exists in maternal plasma (≥ 10%) at as early as 6 weeks of gestation; the concentration rises during pregnancy and peaks prior to parturition [4]. Jorgez described enrichment of cff DNA by size fractionation [4]. They showed that 50% of the cell-free DNA in the 100-300- bp fraction separated by agarose gel electrophoresis was of fetal origin, and 30% in the 500-700-bp fraction was of fetal origin. This size-dependent cff DNA distribution is important, possibly making it easier to investigate fetal status (methylome).
The author has no conflict of interest to declare.
The author thanks to Drs. Toshitaka Tanaka and Naho Ikeda for sample collection.


