Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Deepak Saravanan and Monisha Mohan*
Received: December 25, 2022; Published: January 25, 2023
*Corresponding author: Monisha Mohan, Department of Science and Humanities, Indian Institute of Information Technology, Design and Manufacturing, Kancheepuram, India
DOI: 10.26717/BJSTR.2023.48.007627
MKK4, a Ser/Thr protein kinase, is one of the critical activators of MAP kinases in response to various mitogenic stimuli. MKK4 induces skin tumorigenesis through oncogenic activation of the ras gene. The MKK4 acts as the main component of JNK activation which directly induces the RAS signaling pathway. Overexpression of MKK4 is associated with the aggressiveness of various tumors including non-small cell lung, breast, prostate, stomach, and colon adenocarcinomas. Downstream signaling of MKK4 is important for the formation of tumors and hence acts as a potential drug target for cancer therapy. Interestingly, in the present study, it was found that Nilotinib and Dacomitinib bind to the catalytic pocket of MKK4 with the affinity of -9.47 and -9.00 kcal/mol respectively. It was observed that Nilotinib interacts with active site residues of MKK4 namely, Arg110, Lys187, Asp247, Ile108, and Leu236. In addition, it was also found that Dacomitinib interacts with active sites namely, Met181, Ser184, Lys187, Ile108, Lys118, Leu180, Leu236, and Thr391. Furthermore, the binding affinity of Dacomitinib and Nilotinib was found to be higher than the reference compound N-(pyridine-3-yl) pyrrolo[2,1-f] [1,2,4] triazin-4-amine. Taken together, our insilico screening data identifies MKK4 inhibitors that would significantly increase the therapeutic effects of MKK4-associated cancers.
Keywords: Non Melanoma Skin Cancer; Molecular Docking; Molecular Dynamics Simulation; Kinase Inhibitors
Abbreviations: NMSC: Non-Melanoma Skin Cancer; THIF: Trihydroxyisoflavone; PLIP: Protein- Ligand Interaction Profiler; MD: Molecular Dynamic; UAMS: University of Arkansas for Medical Sciences; RMSF: Root-Mean-Square Fluctuation; RMSD: Root-Mean-Square Deviation; HBs: Hydrogen Bonds; RG: Radius of Gyration; SASA: Solvent Accessible Surface Area
Malignant skin cancers are the most dreadful human neoplasms that pose severe medical and social problems to the healthcare sector globally. Skin tumors are broadly classified into two major categories namely; melanoma and non-melanoma skin cancer (NMSC). The development of skin malignancies involves multiple pathways that are triggered by various genetic, environmental factors, diet, and other factors. Exposure to UV radiation is a major etiologic factor that induces photocarcinogenesis leading to inflammation and dysregulation of cellular pathways. Till date, there are no FDAapproved drugs available that would lead to complete regression of the skin tumor. MKK4, a multi-faceted kinase, is one of the key players in the MAPK signaling cascade. MKK4 phosphorylates p38 and JNKs which leads to the activation of the eukaryotic transcription factor NF-κB. Therefore, by boosting the JNK signaling pathway, MKK4 stimulates cell proliferation and the growth of malignancies [1,2].
Previously, it was reported that there is an overexpression of MKK4 in colon and stomach adenocarcinomas, but not in the normal tissues. MKK4 has a non-redundant role in the activation of p38 and JNKs by UV irradiation-stimulated skin tumors [3]. Therefore, targeting MKK4 which is one of the important upstream molecules of the p38 and JNK pathway would be a promising chemoprevention approach for the therapy of skin tumorigenesis. The structure of MKK4 has a kinase fold with 2 lobes namely, a smaller N-terminal and a larger C-terminal lobe which is linked by a flexible hinge region [4]. The ATP binding pocket of MKK4 is located in the cleft between the two lobes. Structural studies have revealed that the residues namely; Glu179 and Met181 in the hinge region form 2 hydrogen bonds with ATP. It was previously reported that ATP-competitive inhibitor 7,3′,4′-trihydroxyisoflavone (THIF) of MKK4, suppresses the UVinduced COX-2 expression thereby reducing the progression of skin cancer [5]. Using the computational approach, the present study has identified potential kinase inhibitors that would block the kinase activity of the MKK4.
Preparation of Protein Target MKK4
The protein structure of MKK4 (PDB: 3ALO) was retrieved from the protein data bank. The PDB consists of structure factor files, coordinate data, and NMR constraint files. Protein structure was prepared using the Pymol software. The pymol software enables us to view the retrieved protein in 3D structure, then it removes the water molecule and adds polar hydrogen and saves it as the pdb format [6]. The MKK4 structure was ionized using EPIK at pH±2 and energy minimized using the OPLS 2005 force field of the Schrodinger suite.
Preparation of Kinase Inhibitors
The kinase inhibitor of about 58 was retrieved from the NCBIPubChem database. The ligand molecules ionized using EPIK at pH±2 and energy minimized using the OPLS 2005 force field of the Schrodinger suite.
Molecular Docking of MKK4 with Kinase Inhibitors
The docking of MKK4 with kinase inhibitors was performed by the SWISSDOCK server. It is the online server based on the EADock DSS engine with the CHARMM22 force field that uses multiple scoring functions [7]. The 3D structure of the docked complex was visualized using the Pymol software [8]. The protein-ligand interaction profiler (PLIP) is an online server which is used for detecting and analyzing the non-covalent interaction between such as hydrogen bonds, salt bridges, hydrophobic and π-cation interactions between the ligand and the receptor. [9,10].
Molecular Dynamic (MD) Simulation
The protein-ligand complex was subjected to Molecular Dynamic (MD) simulations by CABS-flex ver 2.0 and WebGRO Macromolecular Simulations server. WebGRO is an online software for academic purposes that was developed by the University of Arkansas for Medical Sciences (UAMS). The topology of the ligand for simulation in WebGRO was performed using GlycoBioChem PRODRG2. The docked protein-ligand complexes were subjected to GROMOS96 43a1 force field for the molecular dynamic simulations. The solvent model used for the docked complexes is SPC (triclinic water box with size 50 × 75 × 70 Å). The sodium chloride ions were added for the neutralization of the system. The steepest descent algorithm (5000 steps) was used for the minimization of the system [11]. The parameters for the MD were 1 bar pressure, number of frames per simulation (1000), time of simulations (100ns) and temperature of 300 K [12]. The molecular simulation by the WebGRO generates root-mean-square fluctuation (RMSF), root-mean-square deviation (RMSD), radius of gyration (Rg), hydrogen bonds (HBs), and solvent accessible surface area (SASA) to estimate the structure stability and the conformation of changes at 300K. The structure flexibility (RMSF) of the docked MKK4 complexes was also evaluated using CABS-flex 2.0) [13]. It is one of the fastest methods for the molecular simulation of protein folding using a simulation engine (CABS coarse-grained model) [14].
Molecular Docking Analysis of Kinase Inhibitors with MKK4
Figure 1 Interaction analysis of Nilotinib antibiotics drug in the active site pocket of MKK4 protein. The structure of MKK4 and Nilotinib are represented in blue and pink sticks. Amino acid residues of MKK4 protein namely Arg110, Asp247, Lys187, Leu236, Ile236 interact with MKK4.
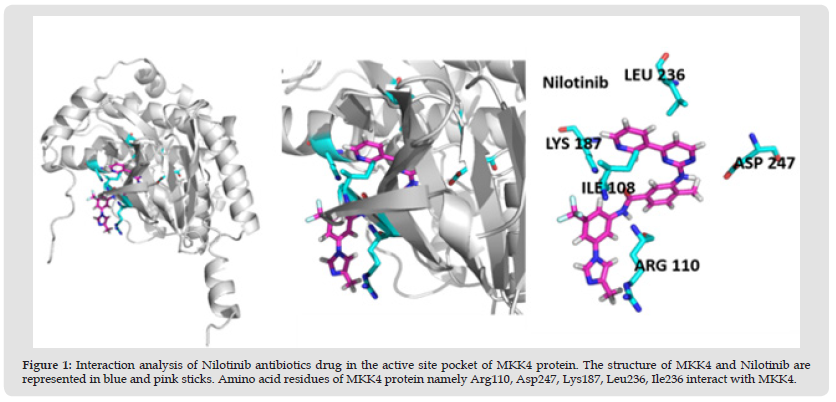
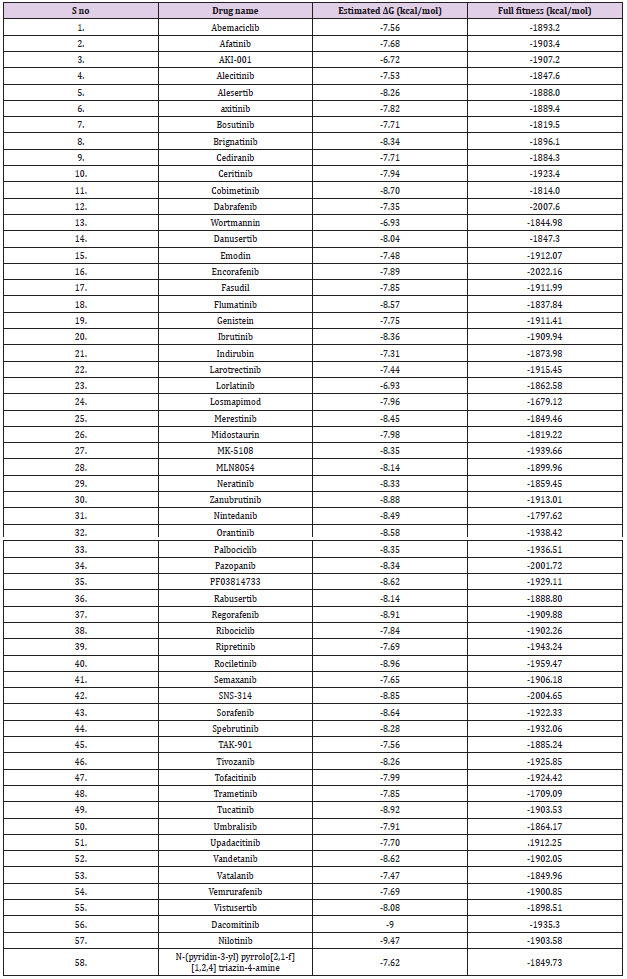
The oncogenic MKK4 triggers tumorigenesis by the activation of the JNK signaling cascade. MKK4 phosphorylates JNKs and p38 kinases on Thr and Tyr residues in the activation loop. The activated JNKs and p38 biological triggers uncontrolled cellular proliferation, cell differentiation, and metastases in various cancers including advanced prostate, skin, stomach, colon, and ovarian cancer [15,16]. MKK4 consists of 399 AA residues which were encoded by the MAP2K4 gene. MKK4 has a typical kinase fold with ATP binding pocket located between the N-terminal and C-terminal lobes [4]. In this study, the ability of 59 kinase inhibitors to block the ATP binding pocket of MKK4 was evaluated. The screening was performed using swissdock to identify the docking conformation of 59 ligands with MKK4 based on the estimated free energy (ΔG). From the molecular docking analysis obtained from the Swissdock, it was found out the two kinase inhibitors namely, Nilotinib and Dacomitinib exhibit the lowest ΔG of -9.47 kcal/mol and -9.00 kcal/mol, respectively (Table 1) (Figure 1). Furthermore, it was observed that Nilotinib and Dacomitinib bind MKK4 with higher affinity in comparison with the reference molecule, N-(pyridine-3-yl)pyrrolo[2,1-f][1,2,4]triazin-4- amine with ΔG of -7.62 kcal/mol.
Table 1: Screening of MKK4 inhibitors derived from antibiotics drugs using Swiss dock software. The compounds Nilotinib and Dacomitinib bind MKK4 with higher affinity of kcal/mol, respectively the binding affinity of the molecule is based on the effective hydrophobic and hydrogen interactions present between the protein and ligand.
Interaction Analysis of MKK4 with Nilotinib and Dacomitinib
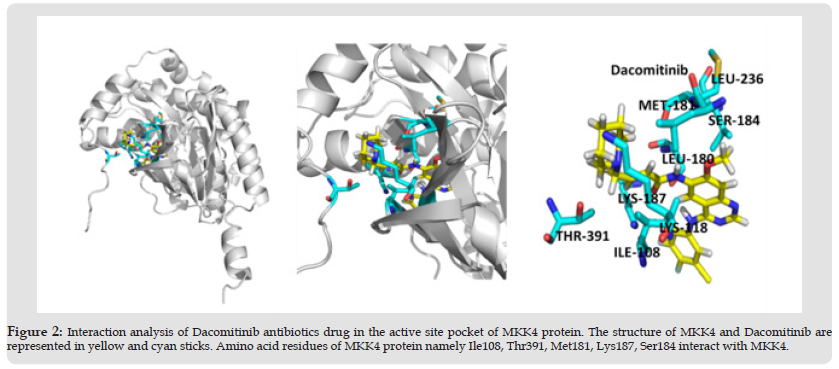
The activation segment of MKK4 consists of amino acid residues Ser257 and Thr261 which are crucial for the phosphorylation of JNK [17]. The role of MKK4 in skin cancer was studied and stated that the oncogenic effect of Ras is essential for the formation of tumors. It was also reported that MKK4 residues namely, Arg 110, Lys 131Asp 185, Ser 184, Lys 187, and Leu 236 are critical for the kinase activity of the MKK4 [18]. Interestingly, the interaction analysis of the MKK4-Nilotinib complex (Table 2), revealed that Nilotinib forms hydrogen bonds with crucial residues namely, Arg110, Lys187, and Asp247, whereas Ile108 and Leu236 are involved in the hydrophobic interaction. The ATP molecule forms hydrogen bonds with Glu179 and Met181 of MKK4 [19]. Surprisingly, the interaction analysis of MKK4 with Dacomitinib indicated that the residues namely, Met181, Ser184, and Lys187 form hydrogen bonds whereas Ile108, Lys118, Leu180, Leu236, and Thr391 form hydrophobic interaction (Table 3) (Figure 2).
Note: Interaction analysis of Nilotinib with MKK4 gives the interacting active binding site residues of Nilotinib namely Arg110, Lys187, Asp247 forms hydrogen interactions. The residues namely Ile108, Leu236 form hydrophobic interaction.
Note: Interaction analysis of Dacomitinib with MKK4 gives the interactive active binding site residues of Dacomitinib namely Met181, Ser184, and Lys187 form hydrogen interactions. The residues namely Ile108, Lys118, Leu180, Leu236, and Thr391 form hydrophobic interaction.
Figure 2 Interaction analysis of Dacomitinib antibiotics drug in the active site pocket of MKK4 protein. The structure of MKK4 and Dacomitinib are represented in yellow and cyan sticks. Amino acid residues of MKK4 protein namely Ile108, Thr391, Met181, Lys187, Ser184 interact with MKK4.
Molecular Dynamic (MD) Simulation of Ligand-MKK4 Complex
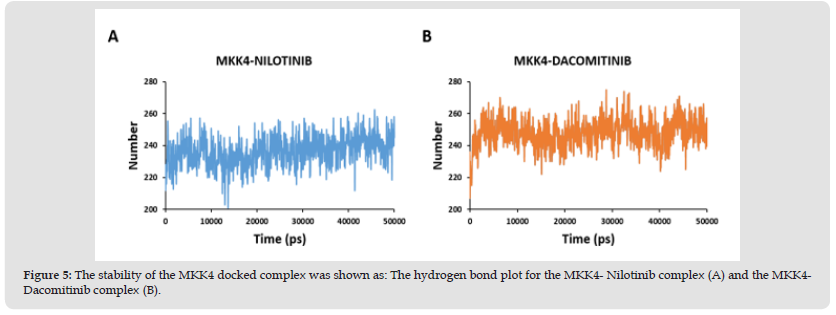
The stability of the protein-ligand complexes was evaluated by molecular dynamics simulation using WebGRO. It detects the alteration in the structure of the protein induced by the ligand during docking. The RMSD value for the protein-ligand less than 2Å is considered to be a stable complex. The RMSD plot of the MKK4- Nilotinib complex demonstrates a stable profile after 10ns with the maximum RMSD value at 0.55 Å (Figure 3A). Furthermore, the MKK4-Dacomitinib complex exhibits the maximum RMSD at 0.67 Å (Figure 3B) achieves a stable profile after the 25ns. RMSF analysis is used to understand the flexibility and stability of the complexes. It determines the changes in the behavior of the amino acid residue of protein while binding with the ligand. The interaction of MKK4- Nilotinib shows moderate flexibility in the residues namely, Ala112 [3.72 Å], Arg262 [3.8 Å], Ser278 [3.43 Å], Trp310 [3.5 Å], and Ala390 [3.24 Å] (Figure 3C) while the residues involved in the interaction namely Arg110, Lys187, Asp247, Ile108, and Leu236 are quite stable. The MKK4-Dacomitinib shows the highest flexibility in the residues namely, Ala112 [4.3 Å], Asp263 [3.44 Å], Gly282 [3.5 Å], and Ala390 [4.01 Å] (Figure 3D) while the interaction residues namely Met181, Ser184, Lys187, Ile108, Lys118, Leu180, Leu236, and Thr391 are quite stable. In addition, CABS-flex was used to determine the flexibility of protein residues (Figure 4). The highest fluctuation for MKK4-Nilotinib and MKK4-Dacominitib are for the residues Arg262 [3.8 Å] and Ala112 [4.3 Å], respectively. Altogether, the RMSF profiles imply that there is a minimum fluctuation of residues located in the ATP binding pocket of MKK4 in complex with the Nilotinib and Dacominitib. The radius of gyration (Rg) is measured to analyze the RMS distance from the rotation axis of the protein. The strength of the protein structure was given by the RG values. The higher Rg values resemble the less compactness of the protein. The interaction of both MKK4-Nilotinib and MKK4-Dacomitinib shows the decreasing Rg value of the backbone till 5ns and there is not much fluctuation between 5-50ns (Figures 3E & 3F). The complex stability depends on the number of hydrogen bonds formed by the ligands with the residues of the ATP binding pocket of MKK4. site of the protein. The length of the hydrogen bonds also plays a major role, shorter length increases the stability. The hydrogen bond plot of the MKK4-Nilotinib complex (Figure 5A) and MKK4-Dacomitinib complex (Figure 5B) shows that residues 260 and 270 form three and five hydrogen bonds respectively. The MKK4-Nilotinib has three hydrogen bonds whereas the MKK4-Dacomitinib has five hydrogen bonds. In conclusion, the RMSF, RMSD, Rg, and Hydrogen plots indicate that the binding of Nilotinib and Dacomitinib does not alter the structure and exhibits a stable association with MKK4 .
Figure 3 The molecular dynamic simulation for the interpretation of stability of the MKK4 given by the following plot: A. The RMSD plot for the MKK4-Nilotinib and B. MKK4-Dacomitinib. C. The RMSF plot for the MKK4-Nilotinib and D. MKK4-Dacomitinib. E. The Rg profiles for the MKK4-Nilotinib and F. MKK4-Dacomitinib.
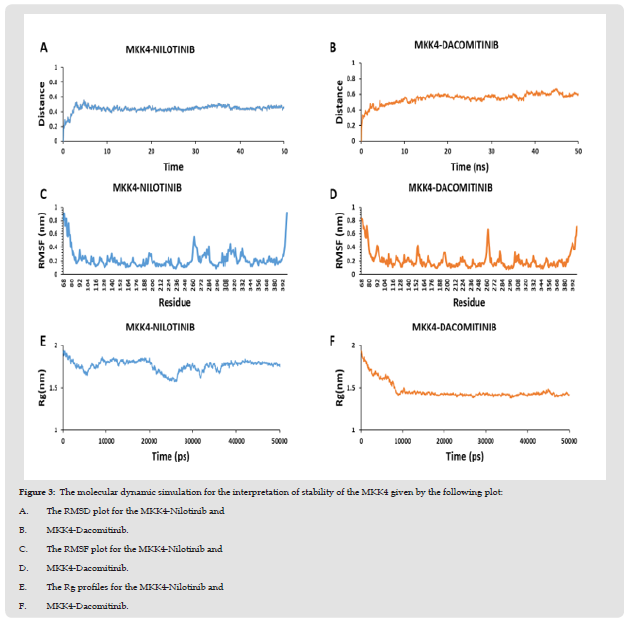
Figure 4 The RMSF plot obtained from the CABS flex 2.0 shows the flexibility of the residue in the receptor for MKK4- Nilotinib complex (A) and the MKK4- Dacomitinib complex (B).
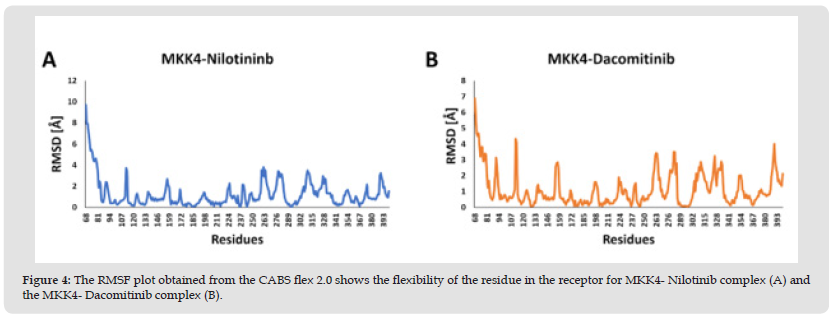
Figure 5 The stability of the MKK4 docked complex was shown as: The hydrogen bond plot for the MKK4- Nilotinib complex (A) and the MKK4- Dacomitinib complex (B).
In recent years, drug discovery using in silico approach has increased drastically as they reduce the cost and time for targeted drug discovery. The number of cases of skin cancer is increasing rapidly due to various factors such as exposure to UV radiation. MKK4 is one of the critical factors that promote skin tumorigenesis in the skin by activating the JNK pathway. Studies have reported that overexpression of MKK4 in tumor tissues is associated with a significantly shorter survival rate. This implies the urgent need for future studies aimed at targeting MKK4 to prevent the progression of malignant skin tumors. In this study, molecular docking and simulation approaches were used to identify a potential kinase inhibitor of MKK4 for targeting skin cancer. Molecular docking analysis revealed that two kinase inhibitors namely, Nilotinib (-9.47 kcal/mol) and Dacomitinib (-9.00 kcal/mol) exhibited the lowest binding energy. To further assess the stability of MKK4-ligand complexes molecular dynamics simulations were performed. The RMSD, RMSF, Rg, and hydrogen plots demonstrate the stable association of Nilotinib and Dacomitinib with MKK4 protein. In conclusion, the efficacy of the inhibitors namely Nilotinib and Dacomitinib could be further validated as a valuable drug for cancer therapy.
We sincerely thank the Indian Institute of Information Technology, Design and Manufacturing – Kancheepuram for providing the necessary infrastructure to carry out this work successfully


